2LOK
 
 | | Solution NMR Structure of the uncharacterized protein from gene locus VNG_0733H of Halobacterium salinarium, Northeast Structural Genomics Consortium Target HsR50 | | Descriptor: | Uncharacterized protein | | Authors: | Rossi, P, Lange, O.F, Lee, H, Hamilton, K, Ciccosanti, C, Buchwald, W.A, Wang, H, Acton, T.B, Xiao, R, Everett, J.K, Montelione, G.T, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2012-01-24 | | Release date: | 2012-02-01 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Determination of solution structures of proteins up to 40 kDa using CS-Rosetta with sparse NMR data from deuterated samples.
Proc.Natl.Acad.Sci.USA, 109, 2012
|
|
2KZN
 
 | | Solution NMR Structure of Peptide methionine sulfoxide reductase msrB from Bacillus subtilis, Northeast Structural Genomics Consortium Target SR10 | | Descriptor: | Peptide methionine sulfoxide reductase msrB | | Authors: | Ertekin, A, Maglaqui, M, Janjua, H, Cooper, B, Ciccosanti, C, Rost, B, Acton, T.B, Xiao, R, Everett, J.K, Prestegard, J, Lee, H, Aramini, J.M, Rossi, P, Montelione, G.T, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2010-06-18 | | Release date: | 2010-07-07 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Determination of solution structures of proteins up to 40 kDa using CS-Rosetta with sparse NMR data from deuterated samples.
Proc.Natl.Acad.Sci.USA, 109, 2012
|
|
2LNA
 
 | | Solution NMR Structure of the mitochondrial inner membrane domain (residues 164-251), FtsH_ext, from the paraplegin-like protein AFG3L2 from Homo sapiens, Northeast Structural Genomics Consortium Target HR6741A | | Descriptor: | AFG3-like protein 2 | | Authors: | Ramelot, T.A, Yang, Y, Lee, H, Janua, H, Kohan, E, Shastry, R, Acton, T.B, Xiao, R, Everett, J.K, Prestegard, J.H, Montelione, G.T, Kennedy, M.A, Northeast Structural Genomics Consortium (NESG), Mitochondrial Protein Partnership (MPP) | | Deposit date: | 2011-12-20 | | Release date: | 2012-01-25 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | NMR structure and MD simulations of the AAA protease intermembrane space domain indicates peripheral membrane localization within the hexaoligomer.
Febs Lett., 587, 2013
|
|
2KY9
 
 | | Solution NMR Structure of ydhK C-terminal Domain from B.subtilis, Northeast Structural Genomics Consortium Target Target SR518 | | Descriptor: | Uncharacterized protein ydhK | | Authors: | Eletsky, A, Sukumaran, D.K, Lee, H, Lee, D, Ciccosanti, C, Janjua, H, Liu, J, Rost, B, Acton, T.B, Xiao, R, Everett, J.K, Prestegard, J.H, Montelione, G.T, Szyperski, T, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2010-05-21 | | Release date: | 2010-07-21 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | The copBL operon protects Staphylococcus aureus from copper toxicity: CopL is an extracellular membrane-associated copper-binding protein.
J.Biol.Chem., 294, 2019
|
|
2KW2
 
 | | Solution NMR of the specialized acyl carrier protein (RPA2022) from Rhodopseudomonas palustris, Northeast Structural Genomics Consortium Target RpR324 | | Descriptor: | Specialized acyl carrier protein | | Authors: | Rossi, P, Lee, H, Wohlbold, T.J, Valafar, H, Lemak, A, Ertekin, A, Forouhar, F, Ciccosanti, C, Rost, B, Acton, T, Xiao, R, Everett, J, Montelione, G.T, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2010-03-30 | | Release date: | 2010-04-21 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Structure of a specialized acyl carrier protein essential for lipid A biosynthesis with very long-chain fatty acids in open and closed conformations.
Biochemistry, 51, 2012
|
|
5FD3
 
 | | Structure of Lin54 tesmin domain bound to DNA | | Descriptor: | DNA (5'-D(*CP*AP*GP*TP*TP*TP*CP*AP*AP*AP*CP*TP*C)-3'), DNA (5'-D(*GP*AP*GP*TP*TP*TP*GP*AP*AP*AP*CP*T)-3'), Protein lin-54 homolog, ... | | Authors: | Marceau, A.H, Felthousen, J.G, Goetsch, P.D, Lee, H, Tripathi, S.M, Strome, S, Litovchick, L, Rubin, S.M. | | Deposit date: | 2015-12-15 | | Release date: | 2016-08-03 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.42 Å) | | Cite: | Structural basis for LIN54 recognition of CHR elements in cell cycle-regulated promoters.
Nat Commun, 7, 2016
|
|
3J8V
 
 | |
3JCX
 
 | | Canine Parvovirus complexed with Fab E | | Descriptor: | Capsid protein 2, Fab E heavy chain, Fab E light chain | | Authors: | Organtini, L.J, Iketani, S, Huang, K, Ashley, R.E, Makhov, A.M, Conway, J.F, Parrish, C.R, Hafenstein, S. | | Deposit date: | 2016-03-21 | | Release date: | 2016-07-20 | | Last modified: | 2018-07-18 | | Method: | ELECTRON MICROSCOPY (4.1 Å) | | Cite: | Near-Atomic Resolution Structure of a Highly Neutralizing Fab Bound to Canine Parvovirus.
J.Virol., 90, 2016
|
|
3J8Z
 
 | |
3J8W
 
 | |
8DSD
 
 | | Human NAMPT in complex with substrate NAM and small molecule activator NP-A1-S | | Descriptor: | (3S)-1-[2-(4-methylphenyl)-2H-pyrazolo[3,4-d]pyrimidin-4-yl]-N-{[4-(methylsulfanyl)phenyl]methyl}piperidine-3-carboxamide, CHLORIDE ION, GLYCEROL, ... | | Authors: | Ratia, K, Xiong, R, Shen, Z, Thatcher, G.R. | | Deposit date: | 2022-07-22 | | Release date: | 2023-03-08 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.429 Å) | | Cite: | Mechanism of Allosteric Modulation of Nicotinamide Phosphoribosyltransferase to Elevate Cellular NAD.
Biochemistry, 62, 2023
|
|
8DSI
 
 | | Human NAMPT in complex with substrate NAM | | Descriptor: | CHLORIDE ION, GLYCEROL, NICOTINAMIDE, ... | | Authors: | Ratia, K, Xiong, R, Shen, Z, Thatcher, G.R. | | Deposit date: | 2022-07-22 | | Release date: | 2023-03-08 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.43 Å) | | Cite: | Mechanism of Allosteric Modulation of Nicotinamide Phosphoribosyltransferase to Elevate Cellular NAD.
Biochemistry, 62, 2023
|
|
8DSH
 
 | | Human NAMPT in complex with quercitrin and AMPcP | | Descriptor: | 2-(3,4-dihydroxyphenyl)-5,7-dihydroxy-4-oxo-4H-chromen-3-yl 6-deoxy-alpha-L-mannopyranoside, Nicotinamide phosphoribosyltransferase, PHOSPHOMETHYLPHOSPHONIC ACID ADENOSYL ESTER | | Authors: | Ratia, K, Xiong, R, Shen, Z, Thatcher, G.R. | | Deposit date: | 2022-07-22 | | Release date: | 2023-03-08 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Mechanism of Allosteric Modulation of Nicotinamide Phosphoribosyltransferase to Elevate Cellular NAD.
Biochemistry, 62, 2023
|
|
8DTJ
 
 | | Human NAMPT in complex with small molecule activator ZN-43-S | | Descriptor: | (3S)-N-[(1-benzothiophen-5-yl)methyl]-1-{2-[4-(trifluoromethyl)phenyl]-2H-pyrazolo[3,4-d]pyrimidin-4-yl}piperidine-3-carboxamide, CHLORIDE ION, GLYCEROL, ... | | Authors: | Ratia, K, Xiong, R, Shen, Z, Thatcher, G.R. | | Deposit date: | 2022-07-25 | | Release date: | 2023-03-08 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.118 Å) | | Cite: | Mechanism of Allosteric Modulation of Nicotinamide Phosphoribosyltransferase to Elevate Cellular NAD.
Biochemistry, 62, 2023
|
|
8DSC
 
 | | Human NAMPT in complex with substrate NAM and small molecule activator NP-A1-R | | Descriptor: | (3R)-1-[2-(4-methylphenyl)-2H-pyrazolo[3,4-d]pyrimidin-4-yl]-N-{[4-(methylsulfanyl)phenyl]methyl}piperidine-3-carboxamide, CHLORIDE ION, GLYCEROL, ... | | Authors: | Ratia, K, Xiong, R, Shen, Z, Thatcher, G.R. | | Deposit date: | 2022-07-22 | | Release date: | 2023-03-08 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.321 Å) | | Cite: | Mechanism of Allosteric Modulation of Nicotinamide Phosphoribosyltransferase to Elevate Cellular NAD.
Biochemistry, 62, 2023
|
|
8DSE
 
 | | Human NAMPT in complex with substrate NAM and activator quercitrin | | Descriptor: | 2-(3,4-dihydroxyphenyl)-5,7-dihydroxy-4-oxo-4H-chromen-3-yl 6-deoxy-alpha-L-mannopyranoside, CHLORIDE ION, NICOTINAMIDE, ... | | Authors: | Ratia, K, Xiong, R, Shen, Z, Thatcher, G.R. | | Deposit date: | 2022-07-22 | | Release date: | 2023-03-08 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.428 Å) | | Cite: | Mechanism of Allosteric Modulation of Nicotinamide Phosphoribosyltransferase to Elevate Cellular NAD.
Biochemistry, 62, 2023
|
|
6B4C
 
 | | Structure of Viperin from Trichoderma virens | | Descriptor: | CITRATE ANION, SULFATE ION, Viperin | | Authors: | Huang, R.H, Selvadurai, K. | | Deposit date: | 2017-09-26 | | Release date: | 2018-07-25 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.795 Å) | | Cite: | Reconstitution and substrate specificity for isopentenyl pyrophosphate of the antiviral radical SAM enzyme viperin.
J.Biol.Chem., 293, 2018
|
|
8E8Y
 
 | | 9H2 Fab-Sabin poliovirus 2 complex | | Descriptor: | 9H2 Fab heavy chain, 9H2 Fab light chain, Capsid protein VP1, ... | | Authors: | Charnesky, A.J. | | Deposit date: | 2022-08-26 | | Release date: | 2023-06-21 | | Last modified: | 2024-10-09 | | Method: | ELECTRON MICROSCOPY (2.5 Å) | | Cite: | A human monoclonal antibody binds within the poliovirus receptor-binding site to neutralize all three serotypes.
Nat Commun, 14, 2023
|
|
8E8R
 
 | | 9H2 Fab-Sabin poliovirus 3 complex | | Descriptor: | 9H2 Fab heavy chain, 9H2 Fab light chain, Capsid protein VP1, ... | | Authors: | Charnesky, A.J. | | Deposit date: | 2022-08-25 | | Release date: | 2023-06-21 | | Last modified: | 2024-10-16 | | Method: | ELECTRON MICROSCOPY (2.66 Å) | | Cite: | A human monoclonal antibody binds within the poliovirus receptor-binding site to neutralize all three serotypes.
Nat Commun, 14, 2023
|
|
8E8X
 
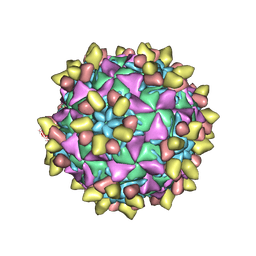 | | 9H2 Fab-Sabin poliovirus 3 complex | | Descriptor: | 9H2 Fab heavy chain, 9H2 Fab light chain, Capsid protein VP1, ... | | Authors: | Charnesky, A.J. | | Deposit date: | 2022-08-26 | | Release date: | 2023-06-21 | | Last modified: | 2023-10-18 | | Method: | ELECTRON MICROSCOPY (2.91 Å) | | Cite: | A human monoclonal antibody binds within the poliovirus receptor-binding site to neutralize all three serotypes.
Nat Commun, 14, 2023
|
|
8E8Z
 
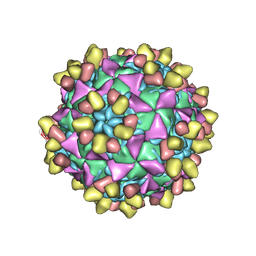 | | 9H2 Fab-Sabin poliovirus 1 complex | | Descriptor: | 9H2 Fab heavy chain, 9H2 Fab light chain, Capsid protein VP1, ... | | Authors: | Charnesky, A.J. | | Deposit date: | 2022-08-26 | | Release date: | 2023-06-21 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (3.15 Å) | | Cite: | A human monoclonal antibody binds within the poliovirus receptor-binding site to neutralize all three serotypes.
Nat Commun, 14, 2023
|
|
8E8S
 
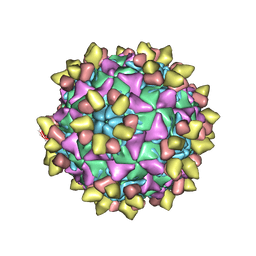 | | 9H2 Fab-poliovirus 2 complex | | Descriptor: | 9H2 Fab heavy chain, 9H2 Fab light chain, Capsid protein VP1, ... | | Authors: | Charnesky, A.J. | | Deposit date: | 2022-08-25 | | Release date: | 2023-06-21 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (2.73 Å) | | Cite: | A human monoclonal antibody binds within the poliovirus receptor-binding site to neutralize all three serotypes.
Nat Commun, 14, 2023
|
|
8E8L
 
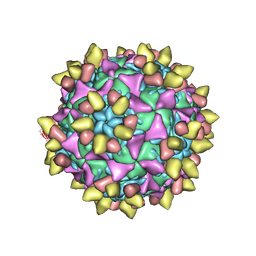 | | 9H2 Fab-poliovirus 1 complex | | Descriptor: | 9H2 Fab heavy chain, 9H2 Fab light chain, Capsid protein VP1, ... | | Authors: | Charnesky, A.J. | | Deposit date: | 2022-08-25 | | Release date: | 2023-06-21 | | Last modified: | 2023-10-18 | | Method: | ELECTRON MICROSCOPY (3.13 Å) | | Cite: | A human monoclonal antibody binds within the poliovirus receptor-binding site to neutralize all three serotypes.
Nat Commun, 14, 2023
|
|
3MJ5
 
 | | Severe Acute Respiratory Syndrome-Coronavirus Papain-Like Protease Inhibitors: Design, Synthesis, Protein-Ligand X-ray Structure and Biological Evaluation | | Descriptor: | N-(1,3-benzodioxol-5-ylmethyl)-1-[(1R)-1-naphthalen-1-ylethyl]piperidine-4-carboxamide, Replicase polyprotein 1a, ZINC ION | | Authors: | Mesecar, A.D, Ratia, K.M, Pegan, S.D. | | Deposit date: | 2010-04-12 | | Release date: | 2010-06-30 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.63 Å) | | Cite: | Severe acute respiratory syndrome coronavirus papain-like novel protease inhibitors: design, synthesis, protein-ligand X-ray structure and biological evaluation
J.Med.Chem., 53, 2010
|
|
7ABU
 
 | | Structure of SARS-CoV-2 Main Protease bound to RS102895 | | Descriptor: | 1'-[2-[4-(trifluoromethyl)phenyl]ethyl]spiro[1~{H}-3,1-benzoxazine-4,4'-piperidine]-2-one, 3C-like proteinase, DIMETHYL SULFOXIDE, ... | | Authors: | Guenther, S, Reinke, P.Y.A, Oberthuer, D, Yefanov, O, Gelisio, L, Ginn, H, Lieske, J, Domaracky, M, Brehm, W, Rahmani Mashour, A, White, T.A, Knoska, J, Pena Esperanza, G, Koua, F, Tolstikova, A, Groessler, M, Fischer, P, Hennicke, V, Fleckenstein, H, Trost, F, Galchenkova, M, Gevorkov, Y, Li, C, Awel, S, Paulraj, L.X, Ullah, N, Falke, S, Alves Franca, B, Schwinzer, M, Brognaro, H, Werner, N, Perbandt, M, Tidow, H, Seychell, B, Beck, T, Meier, S, Doyle, J.J, Giseler, H, Melo, D, Lane, T.J, Dunkel, I, Peck, A, Saouane, S, Hakanpaeae, J, Meyer, J, Noei, H, Gribbon, P, Ellinger, B, Kuzikov, M, Wolf, M, Zhang, L, Ehrt, C, Pletzer-Zelgert, J, Wollenhaupt, J, Feiler, C, Weiss, M, Schulz, E.C, Mehrabi, P, Norton-Baker, B, Schmidt, C, Lorenzen, K, Schubert, R, Han, H, Chari, A, Fernandez Garcia, Y, Turk, D, Hilgenfeld, R, Rarey, M, Zaliani, A, Chapman, H.N, Pearson, A, Betzel, C, Meents, A. | | Deposit date: | 2020-09-08 | | Release date: | 2020-12-02 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | X-ray screening identifies active site and allosteric inhibitors of SARS-CoV-2 main protease.
Science, 372, 2021
|
|
