3HE7
 
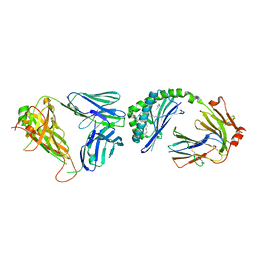 | | Crystal structure of mouse CD1d-alpha-galactosylceramide with mouse Valpha14-Vbeta7 NKT TCR | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Antigen-presenting glycoprotein CD1d1, ... | | Authors: | Patel, O, Rossjohn, J. | | Deposit date: | 2009-05-08 | | Release date: | 2009-07-28 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Differential recognition of CD1d-alpha-galactosyl ceramide by the V beta 8.2 and V beta 7 semi-invariant NKT T cell receptors
Immunity, 31, 2009
|
|
4PJG
 
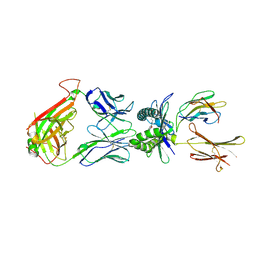 | | Structure of human MR1-Ac-6-FP in complex with human MAIT B-F3-C1 TCR | | Descriptor: | Beta-2-microglobulin, Major histocompatibility complex class I-related gene protein, N-(6-formyl-4-oxo-3,4-dihydropteridin-2-yl)acetamide, ... | | Authors: | Birkinshaw, R.W, Rossjohn, J. | | Deposit date: | 2014-05-12 | | Release date: | 2014-07-02 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | A molecular basis underpinning the T cell receptor heterogeneity of mucosal-associated invariant T cells.
J.Exp.Med., 211, 2014
|
|
4PJX
 
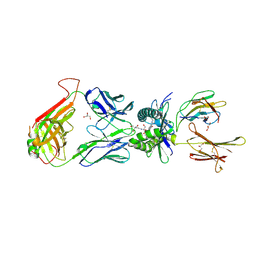 | | Structure of human MR1-Ac-6-FP in complex with human MAIT C-A11 TCR | | Descriptor: | 2-[3-(2-HYDROXY-1,1-DIHYDROXYMETHYL-ETHYLAMINO)-PROPYLAMINO]-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, Beta-2-microglobulin, CHLORIDE ION, ... | | Authors: | Birkinshaw, R.W, Rossjohn, J. | | Deposit date: | 2014-05-12 | | Release date: | 2014-07-02 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | A molecular basis underpinning the T cell receptor heterogeneity of mucosal-associated invariant T cells.
J.Exp.Med., 211, 2014
|
|
4DZB
 
 | |
3SDA
 
 | |
3SDC
 
 | |
2NW3
 
 | | Crystal structure of HLA-B*3508 presenting EBV peptide EPLPQGQLTAY at 1.7A | | Descriptor: | Beta-2-microglobulin, EBV peptide EPLPQGQLTAY, HLA class I histocompatibility antigen, ... | | Authors: | Tynan, F.E, Reid, H.H, Rossjohn, J. | | Deposit date: | 2006-11-14 | | Release date: | 2007-02-27 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | A T cell receptor flattens a bulged antigenic peptide presented by a major histocompatibility complex class I molecule
Nat.Immunol., 8, 2007
|
|
2NW2
 
 | | Crystal structure of ELS4 TCR at 1.4A | | Descriptor: | ELS4 TCR alpha chain, ELS4 TCR beta chain | | Authors: | Tynan, F.E, Reid, H.H, Rossjohn, J. | | Deposit date: | 2006-11-14 | | Release date: | 2007-02-27 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | A T cell receptor flattens a bulged antigenic peptide presented by a major histocompatibility complex class I molecule
Nat.Immunol., 8, 2007
|
|
3SDD
 
 | |
2NX5
 
 | | Crystal structure of ELS4 TCR bound to HLA-B*3501 presenting EBV peptide EPLPQGQLTAY at 1.7A | | Descriptor: | Beta-2-microglobulin, EBV peptide, EPLPQGQLTAY, ... | | Authors: | Tynan, F.E, Reid, H.H, Rossjohn, J. | | Deposit date: | 2006-11-16 | | Release date: | 2007-02-27 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | A T cell receptor flattens a bulged antigenic peptide presented by a major histocompatibility complex class I molecule
Nat.Immunol., 8, 2007
|
|
3SCM
 
 | |
3TO4
 
 | | Structure of mouse Valpha14Vbeta2-mouseCD1d-alpha-Galactosylceramide | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Antigen-presenting glycoprotein CD1d1, ... | | Authors: | Patel, O, Rossjohn, J. | | Deposit date: | 2011-09-04 | | Release date: | 2011-12-14 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Vbeta2 natural killer T cell antigen receptor-mediated recognition of CD1d-glycolipid antigen.
Proc.Natl.Acad.Sci.USA, 108, 2011
|
|
2PO6
 
 | |
2NNA
 
 | | Structure of the MHC class II molecule HLA-DQ8 bound with a deamidated gluten peptide | | Descriptor: | MHC class II antigen, gluten peptide | | Authors: | Henderson, K.N, Tye-Din, J.A, Rossjohn, J, Anderson, R.P. | | Deposit date: | 2006-10-24 | | Release date: | 2007-09-04 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | A structural and immunological basis for the role of human leukocyte antigen DQ8 in celiac disease
Immunity, 27, 2007
|
|
3VFW
 
 | | crystal structure of HLA B*3508 LPEP-P10Ala, peptide mutant P10-ala | | Descriptor: | Beta-2-microglobulin, LPEP peptide from EBV, P10A, ... | | Authors: | Liu, Y.C, Rossjohn, J, Gras, S. | | Deposit date: | 2012-01-10 | | Release date: | 2012-02-22 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | The Energetic Basis Underpinning T-cell Receptor Recognition of a Super-bulged Peptide Bound to a Major Histocompatibility Complex Class I Molecule.
J.Biol.Chem., 287, 2012
|
|
3VFV
 
 | | crystal structure of HLA B*3508 LPEP-P9Ala, peptide mutant P9-ala | | Descriptor: | Beta-2-microglobulin, LPEP peptide from EBV, P9A, ... | | Authors: | Liu, Y.C, Rossjohn, J, Gras, S. | | Deposit date: | 2012-01-10 | | Release date: | 2012-02-22 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | The Energetic Basis Underpinning T-cell Receptor Recognition of a Super-bulged Peptide Bound to a Major Histocompatibility Complex Class I Molecule.
J.Biol.Chem., 287, 2012
|
|
3TN0
 
 | |
6CRJ
 
 | |
3RUG
 
 | | Crystal structure of Valpha10-Vbeta8.1 NKT TCR in complex with CD1d-alphaglucosylceramide (C20:2) | | Descriptor: | (11E,14E)-N-[(2S,3S,4R)-1-(alpha-D-glucopyranosyloxy)-3,4-dihydroxyoctadecan-2-yl]icosa-11,14-dienamide, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Patel, O, Rossjohn, J. | | Deposit date: | 2011-05-05 | | Release date: | 2011-08-03 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | A semi-invariant V(alpha)10(+) T cell antigen receptor defines a population of natural killer T cells with distinct glycolipid antigen-recognition properties
Nat.Immunol., 12, 2011
|
|
3QI9
 
 | | Crystal structure of mouse CD1d-alpha-phosphotidylinositol with mouse Valpha14-Vbeta6 2A3-D NKT TCR | | Descriptor: | 2-[(HYDROXY{[(2R,3R,5S,6R)-2,3,4,5,6-PENTAHYDROXYCYCLOHEXYL]OXY}PHOSPHORYL)OXY]-1-[(PALMITOYLOXY)METHYL]ETHYL HEPTADECANOATE, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Clarke, A.J, Rossjohn, J. | | Deposit date: | 2011-01-26 | | Release date: | 2012-02-15 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | A molecular basis for NKT cell recognition of CD1d-self-antigen
Immunity, 34, 2011
|
|
3TGR
 
 | | Crystal structure of unliganded HIV-1 clade C strain C1086 gp120 core | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, HIV-1 clade C1086 gp120 | | Authors: | Kwon, Y.D, Kwong, P.D. | | Deposit date: | 2011-08-17 | | Release date: | 2012-04-04 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Unliganded HIV-1 gp120 core structures assume the CD4-bound conformation with regulation by quaternary interactions and variable loops.
Proc.Natl.Acad.Sci.USA, 109, 2012
|
|
3TGT
 
 | | Crystal structure of unliganded HIV-1 clade A/E strain 93TH057 gp120 core | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, HIV-1 clade A/E 93TH057 gp120 | | Authors: | Kwon, Y.D, Kwong, P.D. | | Deposit date: | 2011-08-17 | | Release date: | 2012-04-04 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Unliganded HIV-1 gp120 core structures assume the CD4-bound conformation with regulation by quaternary interactions and variable loops.
Proc.Natl.Acad.Sci.USA, 109, 2012
|
|
3TIH
 
 | |
3TGQ
 
 | | Crystal structure of unliganded HIV-1 clade B strain YU2 gp120 core | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, HIV-1 YU2 gp120 | | Authors: | Kwon, Y.D, Kwong, P.D. | | Deposit date: | 2011-08-17 | | Release date: | 2012-04-04 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (3.4 Å) | | Cite: | Unliganded HIV-1 gp120 core structures assume the CD4-bound conformation with regulation by quaternary interactions and variable loops.
Proc.Natl.Acad.Sci.USA, 109, 2012
|
|
3TGS
 
 | | Crystal structure of HIV-1 clade C strain C1086 gp120 core in complex with NBD-556 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, HIV-1 clade C1086 gp120 core, N-(4-chlorophenyl)-N'-(2,2,6,6-tetramethylpiperidin-4-yl)ethanediamide | | Authors: | Kwon, Y.D, Kwong, P.D. | | Deposit date: | 2011-08-17 | | Release date: | 2012-04-04 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Unliganded HIV-1 gp120 core structures assume the CD4-bound conformation with regulation by quaternary interactions and variable loops.
Proc.Natl.Acad.Sci.USA, 109, 2012
|
|
