2IWO
 
 | | 12th PDZ domain of Multiple PDZ Domain Protein MPDZ | | Descriptor: | MULTIPLE PDZ DOMAIN PROTEIN | | Authors: | Elkins, J.M, Yang, X, Gileadi, C, Schoch, G, Johansson, C, Savitsky, P, Berridge, G, Smee, C.E.A, Turnbull, A, Pike, A, Papagrigoriou, E, Sundstrom, M, Edwards, A, Arrowsmith, C, Weigelt, J, Doyle, D.A. | | Deposit date: | 2006-07-03 | | Release date: | 2006-07-26 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structure of Pick1 and Other Pdz Domains Obtained with the Help of Self-Binding C-Terminal Extensions.
Protein Sci., 16, 2007
|
|
2IWP
 
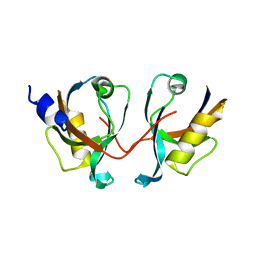 | | 12th PDZ domain of Multiple PDZ Domain Protein MPDZ | | Descriptor: | MULTIPLE PDZ DOMAIN PROTEIN | | Authors: | Elkins, J.M, Yang, X, Gileadi, C, Schoch, G, Johansson, C, Savitsky, P, Berridge, G, Smee, C.E.A, Turnbull, A, Pike, A, Papagrigoriou, E, Sundstrom, M, Edwards, A, Arrowsmith, C, Weigelt, J, Doyle, D.A. | | Deposit date: | 2006-07-03 | | Release date: | 2006-07-26 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Structure of Pick1 and Other Pdz Domains Obtained with the Help of Self-Binding C-Terminal Extension.
Protein Sci., 16, 2007
|
|
4RCA
 
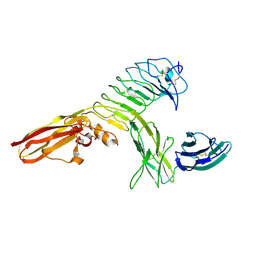 | | Crystal structure of human PTPdelta and human Slitrk1 complex | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Receptor-type tyrosine-protein phosphatase delta, SLIT and NTRK-like protein 1, ... | | Authors: | Kim, H.M, Park, B.S, Kim, D, Lee, S.G. | | Deposit date: | 2014-09-15 | | Release date: | 2014-11-19 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.9908 Å) | | Cite: | Structural basis for LAR-RPTP/Slitrk complex-mediated synaptic adhesion.
Nat Commun, 5, 2014
|
|
8PSE
 
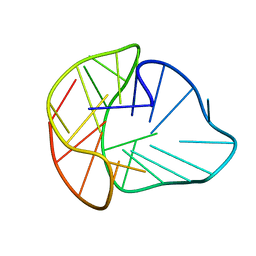 | |
8PSI
 
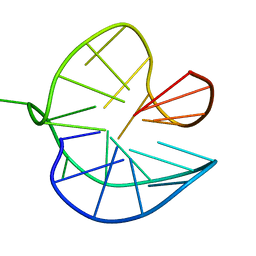 | |
8PSC
 
 | |
8FFR
 
 | | Revised structure of the rabies virus nucleoprotein-RNA complex | | Descriptor: | Nucleoprotein, PHOSPHATE ION, RNA (99-MER) | | Authors: | Leyrat, C, Bourhis, J.M, Albertini, A.A.V, Wernimont, A.K, Muziol, T, Ravelli, R.B.G, Weissenhorn, W, Ruigrok, R.W.H, Jamin, M. | | Deposit date: | 2022-12-09 | | Release date: | 2023-01-11 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (3.49 Å) | | Cite: | Structure and Dynamics of the Unassembled Nucleoprotein of Rabies Virus in Complex with Its Phosphoprotein Chaperone Module.
Viruses, 14, 2022
|
|
1PDI
 
 | | Fitting of the C-terminal part of the short tail fibers into the cryo-EM reconstruction of T4 baseplate | | Descriptor: | Short tail fiber protein | | Authors: | Kostyuchenko, V.A, Leiman, P.G, Chipman, P.R, Kanamaru, S, van Raaij, M.J, Arisaka, F, Mesyanzhinov, V.V, Rossmann, M.G. | | Deposit date: | 2003-05-19 | | Release date: | 2003-09-09 | | Last modified: | 2024-02-14 | | Method: | ELECTRON MICROSCOPY (12 Å) | | Cite: | Three-dimensional structure of bacteriophage T4 baseplate
Nat.Struct.Biol., 10, 2003
|
|
1LRZ
 
 | | x-ray crystal structure of staphylococcus aureus femA | | Descriptor: | factor essential for expression of methicillin resistance | | Authors: | Benson, T, Prince, D, Mutchler, V, Curry, K, Ho, A, Sarver, R, Hagadorn, J, Choi, G, Garlick, R. | | Deposit date: | 2002-05-16 | | Release date: | 2002-09-04 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | X-ray crystal structure of Staphylococcus aureus FemA.
Structure, 10, 2002
|
|
1LMH
 
 | |
5CPR
 
 | | The novel SUV4-20 inhibitor A-196 verifies a role for epigenetics in genomic integrity | | Descriptor: | 6,7-dichloro-N-cyclopentyl-4-(pyridin-4-yl)phthalazin-1-amine, Histone-lysine N-methyltransferase SUV420H1, S-ADENOSYLMETHIONINE, ... | | Authors: | Jakob, C.G, Upadhyay, A.K, Sun, C. | | Deposit date: | 2015-07-21 | | Release date: | 2017-01-25 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.22 Å) | | Cite: | The SUV4-20 inhibitor A-196 verifies a role for epigenetics in genomic integrity.
Nat. Chem. Biol., 13, 2017
|
|
4MIC
 
 | | Crystal structure of Gpb in complex with SUGAR (N-{(2E)-3-[4-(PROPAN-2-YL)PHENYL]PROP-2-ENOYL}-BETA-D-GLUCOPYRANOSYLAMINE) (S6) | | Descriptor: | Glycogen phosphorylase, muscle form, N-{(2E)-3-[4-(propan-2-yl)phenyl]prop-2-enoyl}-beta-D-glucopyranosylamine | | Authors: | Kantsadi, A.L, Chatzileontiadou, D.S.M, Leonidas, D.D. | | Deposit date: | 2013-08-30 | | Release date: | 2014-07-23 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (2.45 Å) | | Cite: | Structure based inhibitor design targeting glycogen phosphorylase b. Virtual screening, synthesis, biochemical and biological assessment of novel N-acyl-beta-d-glucopyranosylamines.
Bioorg.Med.Chem., 22, 2014
|
|
4PB6
 
 | |
2WZN
 
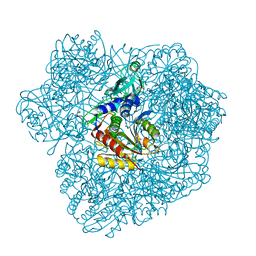 | | 3d structure of TET3 from Pyrococcus horikoshii | | Descriptor: | 354AA LONG HYPOTHETICAL OPERON PROTEIN FRV, CHLORIDE ION, GLYCEROL, ... | | Authors: | Rosenbaum, E, Dura, M.A, Vellieux, F.M, Franzetti, B. | | Deposit date: | 2009-12-01 | | Release date: | 2010-11-03 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | The Structural and Biochemical Characterizations of a Novel Tet Peptidase Complex from Pyrococcus Horikoshii Reveal an Integrated Peptide Degradation System in Hyperthermophilic Archaea.
Mol.Microbiol., 72, 2009
|
|
2WYR
 
 | |
2JM5
 
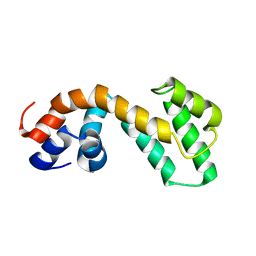 | | Solution Structure of the RGS domain from human RGS18 | | Descriptor: | Regulator of G-protein signaling 18 | | Authors: | Higman, V.A, Leidert, M, Bray, J, Elkins, J, Soundararajan, M, Doyle, D.A, Gileadi, C, Phillips, C, Schoch, G, Yang, X, Brockmann, C, Schmieder, P, Diehl, A, Sundstrom, M, Arrowsmith, C, Weigelt, J, Edwards, A, Oschkinat, H, Ball, L.J, Structural Genomics Consortium (SGC) | | Deposit date: | 2006-10-11 | | Release date: | 2006-10-24 | | Last modified: | 2024-05-08 | | Method: | SOLUTION NMR | | Cite: | Structural diversity in the RGS domain and its interaction with heterotrimeric G protein alpha-subunits.
Proc.Natl.Acad.Sci.Usa, 105, 2008
|
|
4MI9
 
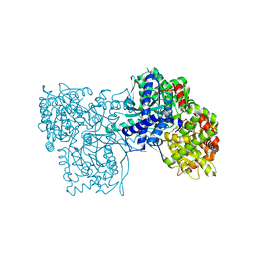 | | Crystal structure of Gpb in complex with SUGAR (N-[(3R)-3-(4-ETHYLPHENYL)BUTANOYL]-BETA-D-GLUCOPYRANOSYLAMINE) (S20) | | Descriptor: | Glycogen phosphorylase, muscle form, N-[(3R)-3-(4-ethylphenyl)butanoyl]-beta-D-glucopyranosylamine | | Authors: | Kantsadi, A.L, Chatzileontiadou, D.S.M, Leonidas, D.D. | | Deposit date: | 2013-08-30 | | Release date: | 2014-07-23 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Structure based inhibitor design targeting glycogen phosphorylase b. Virtual screening, synthesis, biochemical and biological assessment of novel N-acyl-beta-d-glucopyranosylamines.
Bioorg.Med.Chem., 22, 2014
|
|
1LWO
 
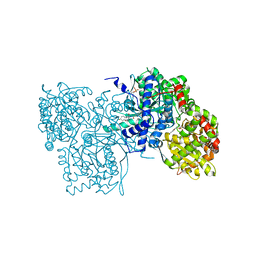 | | Crystal structure of rabbit muscle glycogen phosphorylase a in complex with a potential hypoglycaemic drug at 2.0 A resolution | | Descriptor: | 5-CHLORO-1H-INDOLE-2-CARBOXYLIC ACID [1-(4-FLUOROBENZYL)-2-(4-HYDROXYPIPERIDIN-1YL)-2-OXOETHYL]AMIDE, PYRIDOXAL-5'-PHOSPHATE, alpha-D-glucopyranose, ... | | Authors: | Oikonomakos, N.G, Chrysina, E.D, Kosmopoulou, M.N, Leonidas, D.D. | | Deposit date: | 2002-06-01 | | Release date: | 2002-06-19 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal structure of rabbit muscle glycogen phosphorylase a in complex with a potential hypoglycaemic drug at 2.0 A resolution
BIOCHEM.BIOPHYS.ACTA PROTEINS & PROTEOMICS, 1647, 2003
|
|
3NBX
 
 | |
1LWN
 
 | | Crystal structure of rabbit muscle glycogen phosphorylase a in complex with a potential hypoglycaemic drug at 2.0 A resolution | | Descriptor: | PYRIDOXAL-5'-PHOSPHATE, alpha-D-glucopyranose, glycogen phosphorylase | | Authors: | Oikonomakos, N.G, Chrysina, E.D, Kosmopoulou, M, Leonidas, D.D. | | Deposit date: | 2002-06-01 | | Release date: | 2002-06-19 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal structure of rabbit muscle glycogen phosphorylase a in complex with a potential hypoglycaemic drug at 2.0 A resolution
BIOCHEM.BIOPHYS.ACTA PROTEINS & PROTEOMICS, 1647, 2003
|
|
2XGF
 
 | | Structure of the bacteriophage T4 long tail fibre needle-shaped receptor-binding tip | | Descriptor: | CARBONATE ION, FE (II) ION, LONG TAIL FIBER PROTEIN P37 | | Authors: | Bartual, S.G, Otero, J.M, Garcia-Doval, C, Llamas-Saiz, A.L, Kahn, R, Fox, G.C, van Raaij, M.J. | | Deposit date: | 2010-06-03 | | Release date: | 2010-11-03 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structure of the bacteriophage T4 long tail fiber receptor-binding tip.
Proc. Natl. Acad. Sci. U.S.A., 107, 2010
|
|
