5II0
 
 | |
8CK6
 
 | | Crystal structure of maize CKO/CKX8 in complex with urea-derived inhibitor 2-[(3,5-dichlorophenyl)carbamoylamino]-4-methoxy-benzamide | | Descriptor: | 1,2-ETHANEDIOL, 2-[[3,5-bis(chloranyl)phenyl]carbamoylamino]-4-methoxy-benzamide, FLAVIN-ADENINE DINUCLEOTIDE, ... | | Authors: | Kopecny, D, Briozzo, P, Morera, S. | | Deposit date: | 2023-02-14 | | Release date: | 2024-02-28 | | Last modified: | 2024-09-04 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Cytokinin oxidase/dehydrogenase inhibitors: progress towards agricultural practice.
J.Exp.Bot., 75, 2024
|
|
8CKQ
 
 | | Crystal structure of maize cytokinin oxidase/dehydrogenase 4 (CKO/CKX4) in complex with inhibitor 2-[(3,5-dichlorophenyl)carbamoylamino]benzamide | | Descriptor: | 2-[[3,5-bis(chloranyl)phenyl]carbamoylamino]benzamide, Cytokinin dehydrogenase 4, FLAVIN-ADENINE DINUCLEOTIDE | | Authors: | Kopecny, D, Briozzo, P, Morera, S. | | Deposit date: | 2023-02-16 | | Release date: | 2024-02-28 | | Last modified: | 2024-09-04 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Cytokinin oxidase/dehydrogenase inhibitors: progress towards agricultural practice.
J.Exp.Bot., 75, 2024
|
|
8CKT
 
 | | Crystal structure of maize cytokinin oxidase/dehydrogenase 4 (CKO/CKX4) in complex with inhibitor 2-[(3,5-dichlorophenyl)carbamoylamino]-4-(trifluoromethoxy)benzamide | | Descriptor: | 2-[[3,5-bis(chloranyl)phenyl]carbamoylamino]-4-(trifluoromethyloxy)benzamide, Cytokinin dehydrogenase 4, FLAVIN-ADENINE DINUCLEOTIDE | | Authors: | Kopecny, D, Briozzo, P, Morera, S. | | Deposit date: | 2023-02-16 | | Release date: | 2024-02-28 | | Last modified: | 2024-09-04 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Cytokinin oxidase/dehydrogenase inhibitors: progress towards agricultural practice.
J.Exp.Bot., 75, 2024
|
|
8CLW
 
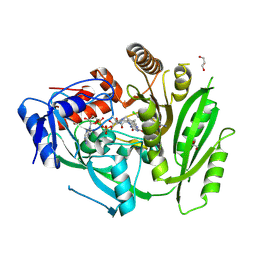 | | Crystal structure of maize cytokinin oxidase/dehydrogenase 4 (CKO/CKX4) in complex with inhibitor 2-[(3,5-dichlorophenyl)carbamoylamino]-4-methoxy-benzamide | | Descriptor: | 1,2-ETHANEDIOL, 2-[[3,5-bis(chloranyl)phenyl]carbamoylamino]-4-methoxy-benzamide, Cytokinin dehydrogenase 4, ... | | Authors: | Kopecny, D, Briozzo, P, Morera, S. | | Deposit date: | 2023-02-17 | | Release date: | 2024-02-28 | | Last modified: | 2024-09-04 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Cytokinin oxidase/dehydrogenase inhibitors: progress towards agricultural practice.
J.Exp.Bot., 75, 2024
|
|
8CM2
 
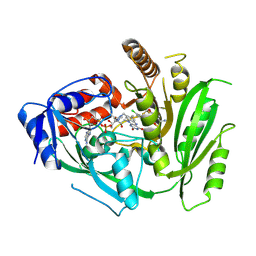 | | Crystal structure of maize cytokinin oxidase/dehydrogenase 4 (CKO/CKX4) in complex with inhibitor 2-[[3,5-dichloro-2-(2-hydroxyethyl)phenyl]carbamoylamino]-4-(trifluoromethoxy)benzamide | | Descriptor: | 2-[[3,5-bis(chloranyl)-2-(2-hydroxyethyl)phenyl]carbamoylamino]-4-(trifluoromethyloxy)benzamide, Cytokinin dehydrogenase 4, FLAVIN-ADENINE DINUCLEOTIDE | | Authors: | Kopecny, D, Briozzo, P, Morera, S. | | Deposit date: | 2023-02-17 | | Release date: | 2024-02-28 | | Last modified: | 2024-09-04 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Cytokinin oxidase/dehydrogenase inhibitors: progress towards agricultural practice.
J.Exp.Bot., 75, 2024
|
|
7OPT
 
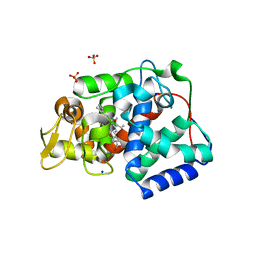 | | Crystal structure of Trypanosoma cruzi peroxidase | | Descriptor: | Ascorbate peroxidase, OXYGEN MOLECULE, PROTOPORPHYRIN IX CONTAINING FE, ... | | Authors: | Freeman, S.L, Kwon, H, Skafar, V, Fielding, A.J, Martinez, A, Piacenza, L, Radi, R, Raven, E.L. | | Deposit date: | 2021-06-01 | | Release date: | 2022-06-08 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.02 Å) | | Cite: | Crystal structure of Trypanosoma cruzi heme peroxidase and characterization of its substrate specificity and compound I intermediate.
J.Biol.Chem., 298, 2022
|
|
7OQR
 
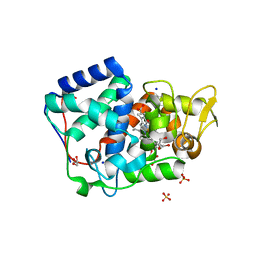 | | Crystal structure of Trypanosoma cruzi peroxidase | | Descriptor: | ACETATE ION, Ascorbate peroxidase, GLYCEROL, ... | | Authors: | Freeman, S.L, Kwon, H, Skafar, V, Fielding, A.J, Martinez, A, Piacenza, L, Radi, R, Raven, E.L. | | Deposit date: | 2021-06-04 | | Release date: | 2022-06-22 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.76 Å) | | Cite: | Crystal structure of Trypanosoma cruzi heme peroxidase and characterization of its substrate specificity and compound I intermediate.
J.Biol.Chem., 298, 2022
|
|
7BGC
 
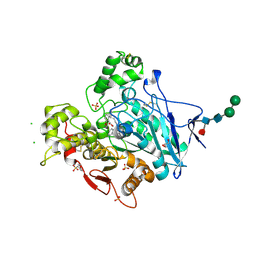 | | human butyrylcholinesterase in complex with a tacrine-methylanacardate hybrid inhibitor | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Nachon, F, Salerno, A, Bolognesi, M.L. | | Deposit date: | 2021-01-06 | | Release date: | 2021-04-28 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Sustainable Drug Discovery of Multi-Target-Directed Ligands for Alzheimer's Disease.
J.Med.Chem., 64, 2021
|
|
7O2O
 
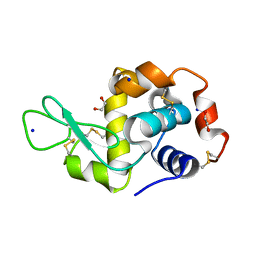 | |
8SZ4
 
 | |
8SZ7
 
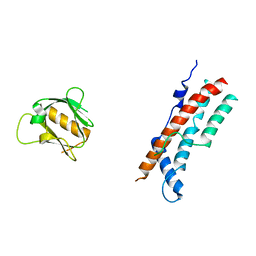 | |
8T0R
 
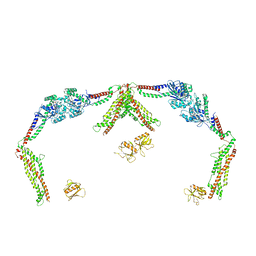 | |
8SXZ
 
 | |
8T0K
 
 | |
8SZ8
 
 | |
8TCD
 
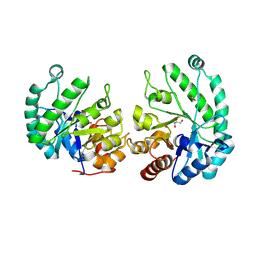 | | Structure of Alistipes sp. 3-Keto-beta-glucopyranoside-1,2-Lyase AL1 | | Descriptor: | ACETATE ION, COBALT (II) ION, GLYCEROL, ... | | Authors: | Lazarski, A.C, Worrall, L.J, Strynadka, N.C.J. | | Deposit date: | 2023-06-30 | | Release date: | 2024-06-12 | | Last modified: | 2024-07-17 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | An alternative broad-specificity pathway for glycan breakdown in bacteria.
Nature, 631, 2024
|
|
8TCS
 
 | | Structure of trehalose bound Alistipes sp. 3-Keto-beta-glucopyranoside-1,2-Lyase AL1 | | Descriptor: | ACETATE ION, COBALT (II) ION, Xylose isomerase-like TIM barrel domain-containing protein, ... | | Authors: | Lazarski, A.C, Worrall, L.J, Strynadka, N.C.J. | | Deposit date: | 2023-07-02 | | Release date: | 2024-06-19 | | Last modified: | 2024-07-17 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | An alternative broad-specificity pathway for glycan breakdown in bacteria.
Nature, 631, 2024
|
|
8TCR
 
 | | Structure of glucose bound Alistipes sp. 3-Keto-beta-glucopyranoside-1,2-Lyase AL1 | | Descriptor: | COBALT (II) ION, MALONATE ION, Sugar phosphate isomerase, ... | | Authors: | Lazarski, A.C, Worrall, L.J, Strynadka, N.C.J. | | Deposit date: | 2023-07-02 | | Release date: | 2024-06-12 | | Last modified: | 2024-07-17 | | Method: | X-RAY DIFFRACTION (2.08 Å) | | Cite: | An alternative broad-specificity pathway for glycan breakdown in bacteria.
Nature, 631, 2024
|
|
6X58
 
 | | MPER-Fluc-Ec2 bound to 10E8v4 antibody | | Descriptor: | 10E8v4 Fab Heavy Chain, 10E8v4 Fab Light Chain, gp41 MPER peptide,Putative fluoride ion transporter CrcB | | Authors: | McIlwain, B.C, Stockbridge, R.B. | | Deposit date: | 2020-05-25 | | Release date: | 2021-05-19 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (3.26 Å) | | Cite: | N-terminal Transmembrane-Helix Epitope Tag for X-ray Crystallography and Electron Microscopy of Small Membrane Proteins.
J.Mol.Biol., 433, 2021
|
|
8TXY
 
 | | X-ray crystal structure of JRD-SIK1/2i-3 bound to a MARK2-SIK2 chimera | | Descriptor: | DI(HYDROXYETHYL)ETHER, N-[(5P,8R)-5-(2-cyano-5-{[(3R)-1-methylpyrrolidin-3-yl]methoxy}pyridin-4-yl)pyrazolo[1,5-a]pyridin-2-yl]cyclopropanecarboxamide, SULFATE ION, ... | | Authors: | Raymond, D.D, Lemke, C.T, Shaffer, P.L, Collins, B, Steele, R, Seierstad, M. | | Deposit date: | 2023-08-24 | | Release date: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Identification of highly selective SIK1/2 inhibitors that modulate innate immune activation and suppress intestinal inflammation.
Proc.Natl.Acad.Sci.USA, 121, 2024
|
|
8TDH
 
 | |
8TYM
 
 | |
8TDA
 
 | |
8TDE
 
 | |
