7XFR
 
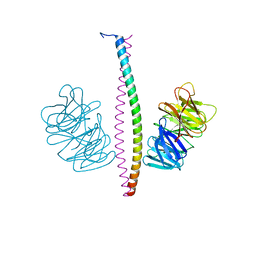 | |
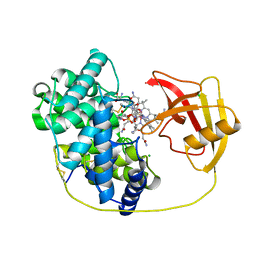
8IXT
 
 | |
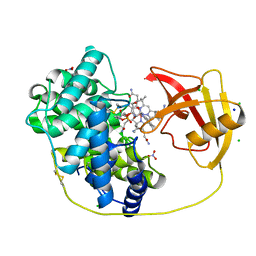
8IXU
 
 | | Rat Transcobalamin in Complex with Cobalamin | | Descriptor: | CHLORIDE ION, COBALAMIN, NITRATE ION, ... | | Authors: | Bokhove, M, Kumasaka, T. | | Deposit date: | 2023-04-03 | | Release date: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | The structure of the rat vitamin B 12 transporter TC and its complex with glutathionylcobalamin.
J.Biol.Chem., 300, 2024
|
|
7YFI
 
 | | Structure of the Rat tri-heteromeric GluN1-GluN2A-GluN2C NMDA receptor in complex with glycine and glutamate | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, GLUTAMIC ACID, ... | | Authors: | Zhang, M, Zhang, J, Guo, F, Li, Y, Zhu, S. | | Deposit date: | 2022-07-08 | | Release date: | 2023-03-29 | | Last modified: | 2023-07-26 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Distinct structure and gating mechanism in diverse NMDA receptors with GluN2C and GluN2D subunits.
Nat.Struct.Mol.Biol., 30, 2023
|
|
7WGV
 
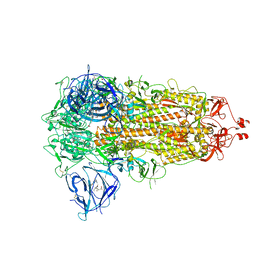 | | SARS-CoV-2 spike glycoprotein trimer in closed state | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 3-[5-[(Z)-(4-ethenyl-3-methyl-5-oxidanylidene-pyrrol-2-ylidene)methyl]-2-[[5-[(Z)-(3-ethenyl-4-methyl-5-oxidanylidene-pyrrol-2-ylidene)methyl]-3-(3-hydroxy-3-oxopropyl)-4-methyl-1H-pyrrol-2-yl]methyl]-4-methyl-1H-pyrrol-3-yl]propanoic acid, ... | | Authors: | Zhu, Y, Tai, L, Yin, G, Sun, F. | | Deposit date: | 2021-12-29 | | Release date: | 2023-01-04 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Novel cleavage sites identified in SARS-CoV-2 spike protein reveal mechanism for cathepsin L-facilitated viral infection and treatment strategies
Cell Discov, 8, 2022
|
|
2Q3X
 
 | | The RIM1alpha C2B domain | | Descriptor: | CHLORIDE ION, Regulating synaptic membrane exocytosis protein 1, SODIUM ION, ... | | Authors: | Guan, R, Dai, H, Tomchick, D.R, Machius, M, Sudhof, T.C, Rizo, J. | | Deposit date: | 2007-05-30 | | Release date: | 2007-08-28 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.73 Å) | | Cite: | Crystal Structure of the RIM1alpha C(2)B Domain at 1.7 A Resolution.
Biochemistry, 46, 2007
|
|
7WG6
 
 | | Neutral Omicron Spike Trimer | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Spike glycoprotein, ... | | Authors: | Cui, Z, Wang, X. | | Deposit date: | 2021-12-28 | | Release date: | 2022-05-18 | | Last modified: | 2022-07-20 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | Structural and functional characterizations of infectivity and immune evasion of SARS-CoV-2 Omicron.
Cell, 185, 2022
|
|
5B2G
 
 | | Crystal structure of human claudin-4 in complex with C-terminal fragment of Clostridium perfringens enterotoxin | | Descriptor: | Endolysin,Claudin-4, Heat-labile enterotoxin B chain | | Authors: | Shinoda, T, Kimura-Someya, T, Shirouzu, M, Yokoyama, S. | | Deposit date: | 2016-01-15 | | Release date: | 2016-10-05 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (3.5 Å) | | Cite: | Structural basis for disruption of claudin assembly in tight junctions by an enterotoxin
Sci Rep, 6, 2016
|
|
7YFG
 
 | | Structure of the Rat GluN1-GluN2C NMDA receptor in complex with glycine and glutamate (major class in asymmetry) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, GLUTAMIC ACID, ... | | Authors: | Zhang, M, Zhang, J, Guo, F, Li, Y, Zhu, S. | | Deposit date: | 2022-07-08 | | Release date: | 2023-03-29 | | Last modified: | 2023-05-31 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | Distinct structure and gating mechanism in diverse NMDA receptors with GluN2C and GluN2D subunits.
Nat.Struct.Mol.Biol., 30, 2023
|
|
2JYJ
 
 | | Re-refining the tetraloop-receptor RNA-RNA complex using NMR-derived restraints and Xplor-nih (2.18) | | Descriptor: | RNA (43-MER) | | Authors: | Zuo, X, Wang, J, Foster, T.R, Schwieters, C.D, Tiede, D.M, Butcher, S.E, Wang, Y. | | Deposit date: | 2007-12-13 | | Release date: | 2008-10-07 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | RNA helical packing in solution: NMR structure of a 30 kDa GAAA tetraloop-receptor complex.
J.Mol.Biol., 351, 2005
|
|
7YFH
 
 | | Structure of the Rat GluN1-GluN2C NMDA receptor in complex with glycine, glutamate and (R)-PYD-106 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, GLUTAMIC ACID, ... | | Authors: | Zhang, M, Zhang, J, Guo, F, Li, Y, Zhu, S. | | Deposit date: | 2022-07-08 | | Release date: | 2023-03-29 | | Last modified: | 2023-05-31 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | Distinct structure and gating mechanism in diverse NMDA receptors with GluN2C and GluN2D subunits.
Nat.Struct.Mol.Biol., 30, 2023
|
|
7YS6
 
 | | Cryo-EM structure of the Serotonin 6 (5-HT6) receptor-DNGs-scFv16 complex | | Descriptor: | 5-hydroxytryptamine receptor 6, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, ... | | Authors: | Zhao, Q.Y, Wang, Y.F, He, L, Wang, S, Cong, Y. | | Deposit date: | 2022-08-11 | | Release date: | 2023-03-29 | | Last modified: | 2023-10-11 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | Structural insights into constitutive activity of 5-HT 6 receptor.
Proc.Natl.Acad.Sci.USA, 120, 2023
|
|
7DWC
 
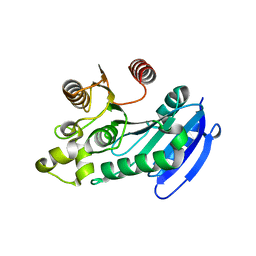 | | Bacteroides thetaiotaomicron VPI5482 BTAxe1 | | Descriptor: | Xylanase | | Authors: | Wang, L.Y, Wang, Y.L, Xin, F.J, Sun, L.C. | | Deposit date: | 2021-01-17 | | Release date: | 2022-01-19 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.804 Å) | | Cite: | Rational Design for Broadened Substrate Specificity and Enhanced Activity of a Novel Acetyl Xylan Esterase from Bacteroides thetaiotaomicron.
J.Agric.Food Chem., 69, 2021
|
|
8PXC
 
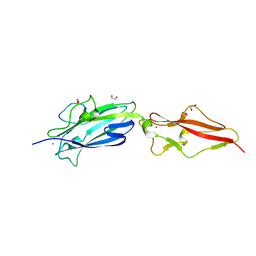 | | Structure of Fap1, a domain of the accessory Sec-dependent serine-rich glycoprotein adhesin from Streptococcus oralis, solved at wavelength 3.06 A | | Descriptor: | 1,2-ETHANEDIOL, CALCIUM ION, Fap1 | | Authors: | El Omari, K, Duman, R, Mykhaylyk, V, Orr, C, Owen, C.D, Walsh, M.A, Wagner, A. | | Deposit date: | 2023-07-23 | | Release date: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.973 Å) | | Cite: | Experimental phasing opportunities for macromolecular crystallography at very long wavelengths.
Commun Chem, 6, 2023
|
|
8PYV
 
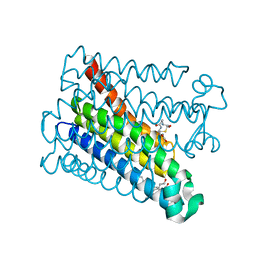 | | Structure of Human PS-1 GSH-analog complex, solved at wavelength 2.755 A | | Descriptor: | L-gamma-glutamyl-S-(2-biphenyl-4-yl-2-oxoethyl)-L-cysteinylglycine, PALMITIC ACID, Prostaglandin E synthase | | Authors: | Duman, R, El Omari, K, Mykhaylyk, V, Orr, C, Wagner, A, Vogeley, L, Brown, D.G. | | Deposit date: | 2023-07-26 | | Release date: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.77 Å) | | Cite: | Experimental phasing opportunities for macromolecular crystallography at very long wavelengths.
Commun Chem, 6, 2023
|
|
8PX9
 
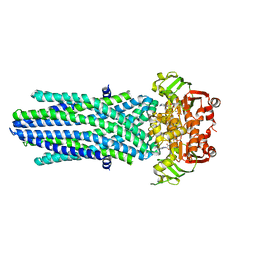 | | Structure of the antibacterial peptide ABC transporter McjD, solved at wavelength 2.75 A | | Descriptor: | MAGNESIUM ION, Microcin-J25 export ATP-binding/permease protein McjD, PHOSPHOAMINOPHOSPHONIC ACID-ADENYLATE ESTER | | Authors: | El Omari, K, Duman, R, Mykhaylyk, V, Orr, C, Bountra, K, Beis, K, Wagner, A. | | Deposit date: | 2023-07-22 | | Release date: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Experimental phasing opportunities for macromolecular crystallography at very long wavelengths.
Commun Chem, 6, 2023
|
|
8PZ4
 
 | | Structure of alginate transporter, AlgE, solved at wavelength 2.755 A | | Descriptor: | (2R)-2,3-DIHYDROXYPROPYL(7Z)-PENTADEC-7-ENOATE, (2S)-2,3-DIHYDROXYPROPYL(7Z)-PENTADEC-7-ENOATE, 3,6,9,12,15,18,21,24-OCTAOXAHEXACOSAN-1-OL, ... | | Authors: | Duman, R, El Omari, K, Mykhaylyk, V, Orr, C, Wagner, A, Vogeley, L, Brown, D.G. | | Deposit date: | 2023-07-26 | | Release date: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.77 Å) | | Cite: | Experimental phasing opportunities for macromolecular crystallography at very long wavelengths.
Commun Chem, 6, 2023
|
|
8PX5
 
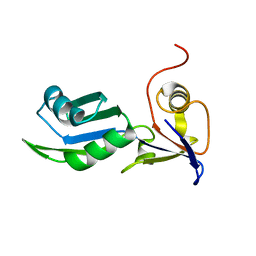 | | Structure of the RNA recognition motif (RRM) of Seb1 from S. pombe., solved at wavelength 2.75 A | | Descriptor: | Rpb7-binding protein seb1 | | Authors: | El Omari, K, Duman, R, Mykhaylyk, V, Orr, C, Wittmann, S, Renner, M, Grimes, J.M, Wagner, A. | | Deposit date: | 2023-07-22 | | Release date: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.77 Å) | | Cite: | Experimental phasing opportunities for macromolecular crystallography at very long wavelengths.
Commun Chem, 6, 2023
|
|
8PX4
 
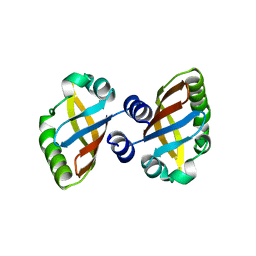 | | Structure of the PAS domain code by the LIC_11128 gene from Leptospira interrogans serovar Copenhageni Fiocruz, solved at wavelength 3.09 A | | Descriptor: | Diguanylate cyclase | | Authors: | El Omari, K, Duman, R, Mykhaylyk, V, Orr, C, Guzzo, C.R, Owens, R.J, Wagner, A. | | Deposit date: | 2023-07-22 | | Release date: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Experimental phasing opportunities for macromolecular crystallography at very long wavelengths.
Commun Chem, 6, 2023
|
|
8PXJ
 
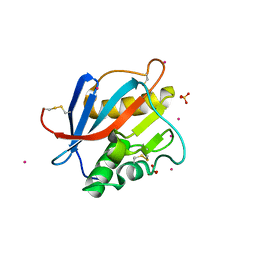 | | Structure of Whitewater Arroyo virus GP1 glycoprotein, solved at wavelength 2.75 A | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, CADMIUM ION, Glycoprotein G1, ... | | Authors: | El Omari, K, Duman, R, Mykhaylyk, V, Orr, C, Bowden, T.A, Wagner, A. | | Deposit date: | 2023-07-23 | | Release date: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.75 Å) | | Cite: | Experimental phasing opportunities for macromolecular crystallography at very long wavelengths.
Commun Chem, 6, 2023
|
|
8PWN
 
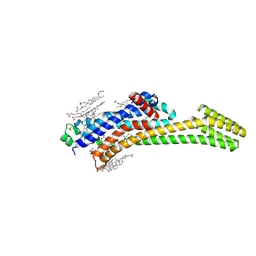 | | Structure of A2A adenosine receptor A2AR-StaR2-bRIL, solved at wavelength 2.75 A | | Descriptor: | Adenosine receptor A2a,Soluble cytochrome b562, CHOLESTEROL, OLEIC ACID, ... | | Authors: | El Omari, K, Duman, R, Mykhaylyk, V, Orr, C, Romano, M, Moraes, I, Wagner, A. | | Deposit date: | 2023-07-20 | | Release date: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Experimental phasing opportunities for macromolecular crystallography at very long wavelengths.
Commun Chem, 6, 2023
|
|
8PX0
 
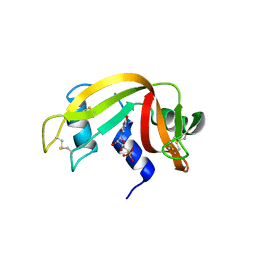 | | Structure of ribonuclease A, solved at wavelength 2.75 A | | Descriptor: | L-URIDINE-5'-MONOPHOSPHATE, Ribonuclease pancreatic | | Authors: | El Omari, K, Duman, R, Mykhaylyk, V, Orr, C, Romano, M, Beis, K, Wagner, A. | | Deposit date: | 2023-07-22 | | Release date: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Experimental phasing opportunities for macromolecular crystallography at very long wavelengths.
Commun Chem, 6, 2023
|
|
8PXG
 
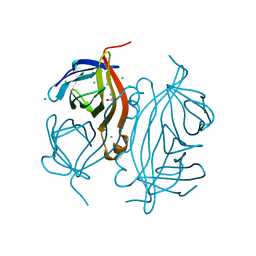 | | Structure of Streptactin, solved at wavelength 2.75 A | | Descriptor: | CHLORIDE ION, GLYCEROL, Streptavidin | | Authors: | El Omari, K, Duman, R, Mykhaylyk, V, Orr, C, Vecchia, L, Jones, E.Y, Wagner, A. | | Deposit date: | 2023-07-23 | | Release date: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Experimental phasing opportunities for macromolecular crystallography at very long wavelengths.
Commun Chem, 6, 2023
|
|
8PX1
 
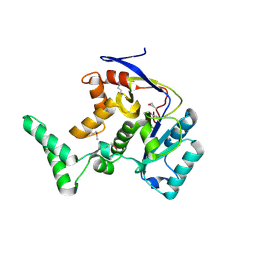 | | Structure of salmonella effector SseK3, solved at wavelength 2.75 A | | Descriptor: | Non-LEE encoded effector protein NleB | | Authors: | El Omari, K, Duman, R, Mykhaylyk, V, Orr, C.M, Esposito, D, Rittinger, K, Wagner, A. | | Deposit date: | 2023-07-22 | | Release date: | 2023-10-25 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Experimental phasing opportunities for macromolecular crystallography at very long wavelengths.
Commun Chem, 6, 2023
|
|
8PXH
 
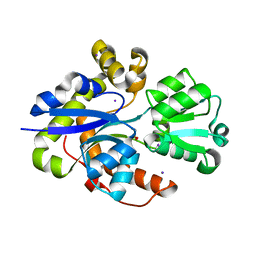 | | Structure of TauA from E. coli, solved at wavelength 2.375 A | | Descriptor: | 2-AMINOETHANESULFONIC ACID, IODIDE ION, Taurine ABC transporter substrate-binding protein | | Authors: | El Omari, K, Duman, R, Mykhaylyk, V, Orr, C, Qu, F, Beis, K, Wagner, A. | | Deposit date: | 2023-07-23 | | Release date: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.77 Å) | | Cite: | Experimental phasing opportunities for macromolecular crystallography at very long wavelengths.
Commun Chem, 6, 2023
|
|
