7XZ6
 
 | | GPR119-Gs-APD668 complex | | Descriptor: | Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, Guanine nucleotide-binding protein G(s) subunit alpha isoforms short, ... | | Authors: | Xu, P, Huang, S, Jiang, Y, Xu, H.E. | | Deposit date: | 2022-06-02 | | Release date: | 2022-08-24 | | Last modified: | 2024-11-20 | | Method: | ELECTRON MICROSCOPY (2.8 Å) | | Cite: | Structural identification of lysophosphatidylcholines as activating ligands for orphan receptor GPR119.
Nat.Struct.Mol.Biol., 29, 2022
|
|
6IUH
 
 | | Crystal structure of GIT1 PBD domain in complex with Liprin-alpha2 | | Descriptor: | ARF GTPase-activating protein GIT1, IODIDE ION, Liprin-alpha-2 | | Authors: | Liang, M, Wei, Z. | | Deposit date: | 2018-11-28 | | Release date: | 2019-02-20 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structural basis of the target-binding mode of the G protein-coupled receptor kinase-interacting protein in the regulation of focal adhesion dynamics.
J. Biol. Chem., 294, 2019
|
|
8GB0
 
 | |
8H4H
 
 | | The apo structure of Aspergillomarasmine A synthetase | | Descriptor: | Rhodanese domain-containing protein | | Authors: | Lu, M, Zhang, J, Wang, Z, Han, L. | | Deposit date: | 2022-10-10 | | Release date: | 2023-10-18 | | Last modified: | 2025-04-30 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Investigation of AMA Synthase from Pyrenophora teres f. teres 0-1 for the Synthesis of Aspergillomarasmine A Analogues and Non-Canonical Amino Acids
Adv.Synth.Catal., 366, 2024
|
|
8H4Q
 
 | | Aspergillomarasmine A biosynthese complex with OPS | | Descriptor: | 2-[({3-HYDROXY-2-METHYL-5-[(PHOSPHONOOXY)METHYL]PYRIDIN-4-YL}METHYL)AMINO]ACRYLIC ACID, Rhodanese domain-containing protein | | Authors: | Lu, M, Zhang, J, Wang, Z, Han, L. | | Deposit date: | 2022-10-11 | | Release date: | 2023-10-18 | | Last modified: | 2025-04-30 | | Method: | X-RAY DIFFRACTION (2.22 Å) | | Cite: | Investigation of AMA Synthase from Pyrenophora teres f. teres 0-1 for the Synthesis of Aspergillomarasmine A Analogues and Non-Canonical Amino Acids
Adv.Synth.Catal., 366, 2024
|
|
6IUI
 
 | | Crystal structure of GIT1 PBD domain in complex with Paxillin LD4 motif | | Descriptor: | ARF GTPase-activating protein GIT1, Paxillin | | Authors: | Liang, M, Wei, Z. | | Deposit date: | 2018-11-28 | | Release date: | 2019-03-06 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Structural basis of the target-binding mode of the G protein-coupled receptor kinase-interacting protein in the regulation of focal adhesion dynamics.
J. Biol. Chem., 294, 2019
|
|
7XAB
 
 | | Crystal structure of PDE4D catalytic domain complexed with compound 22d | | Descriptor: | 9-(cyclopropylmethoxy)-8-methoxy-2,2-dimethyl-7-(3-methylbut-2-enyl)-5-(pyridin-4-ylmethoxy)pyrano[3,2-b]xanthen-6-one, Isoform 3 of cAMP-specific 3',5'-cyclic phosphodiesterase 4D, MAGNESIUM ION, ... | | Authors: | Huang, Y.-Y, He, X, Luo, H.-B. | | Deposit date: | 2022-03-17 | | Release date: | 2023-02-22 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.00067449 Å) | | Cite: | Discovery of novel PDE4 inhibitors targeting the M-pocket from natural mangostanin with improved safety for the treatment of Inflammatory Bowel Diseases.
Eur.J.Med.Chem., 242, 2022
|
|
7XAA
 
 | | Crystal structure of PDE4D catalytic domain complexed with compound 21d | | Descriptor: | 8-methoxy-2,2-dimethyl-7-(3-methylbut-2-enyl)-9-oxidanyl-5-(pyridin-4-ylmethoxy)pyrano[3,2-b]xanthen-6-one, Isoform 3 of cAMP-specific 3',5'-cyclic phosphodiesterase 4D, MAGNESIUM ION, ... | | Authors: | Huang, Y.-Y, He, X, Luo, H.-B. | | Deposit date: | 2022-03-17 | | Release date: | 2023-02-22 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.100414 Å) | | Cite: | Discovery of novel PDE4 inhibitors targeting the M-pocket from natural mangostanin with improved safety for the treatment of Inflammatory Bowel Diseases.
Eur.J.Med.Chem., 242, 2022
|
|
7F2L
 
 | | Crystal structure of PDE4D catalytic domain complexed with compound 18a | | Descriptor: | (~{E})-4-[9-[(4-fluorophenyl)methoxy]-8-methoxy-2,2-dimethyl-7-(3-methylbut-2-enyl)-6-oxidanylidene-pyrano[3,2-b]xanthen-5-yl]oxybut-2-enoic acid, Isoform 3 of cAMP-specific 3',5'-cyclic phosphodiesterase 4D, MAGNESIUM ION, ... | | Authors: | Huang, Y.-Y, He, X, Luo, H.-B. | | Deposit date: | 2021-06-11 | | Release date: | 2021-10-20 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.10111427 Å) | | Cite: | Mangostanin Derivatives as Novel and Orally Active Phosphodiesterase 4 Inhibitors for the Treatment of Idiopathic Pulmonary Fibrosis with Improved Safety.
J.Med.Chem., 64, 2021
|
|
7F2M
 
 | | Crystal structure of PDE4D catalytic domain complexed with compound 18d | | Descriptor: | (~{Z})-4-[9-[(4-fluorophenyl)methoxy]-8-methoxy-2,2-dimethyl-7-(3-methylbut-2-enyl)-6-oxidanylidene-pyrano[3,2-b]xanthen-5-yl]oxybut-2-enoic acid, Isoform 3 of cAMP-specific 3',5'-cyclic phosphodiesterase 4D, MAGNESIUM ION, ... | | Authors: | Huang, Y.-Y, He, X, Luo, H.-B. | | Deposit date: | 2021-06-11 | | Release date: | 2021-10-20 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.20004153 Å) | | Cite: | Mangostanin Derivatives as Novel and Orally Active Phosphodiesterase 4 Inhibitors for the Treatment of Idiopathic Pulmonary Fibrosis with Improved Safety.
J.Med.Chem., 64, 2021
|
|
7F2K
 
 | | Crystal structure of PDE4D catalytic domain complexed with compound 17a | | Descriptor: | (~{E})-4-[8-methoxy-2,2-dimethyl-7-(3-methylbut-2-enyl)-9-oxidanyl-6-oxidanylidene-pyrano[3,2-b]xanthen-5-yl]oxybut-2-enoic acid, Isoform 3 of cAMP-specific 3',5'-cyclic phosphodiesterase 4D, MAGNESIUM ION, ... | | Authors: | Huang, Y.-Y, He, X, Luo, H.-B. | | Deposit date: | 2021-06-11 | | Release date: | 2021-10-20 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.10001969 Å) | | Cite: | Mangostanin Derivatives as Novel and Orally Active Phosphodiesterase 4 Inhibitors for the Treatment of Idiopathic Pulmonary Fibrosis with Improved Safety.
J.Med.Chem., 64, 2021
|
|
7WJ6
 
 | |
7WJ7
 
 | |
7END
 
 | |
7EN9
 
 | | Crystal structure of SARS-CoV-2 3CLpro in complex with the non-covalent inhibitor WU-02 | | Descriptor: | 3C-like proteinase, 5-bromanyl-~{N}-methyl-3-nitro-2-[(4~{R},5~{S})-2-(7-oxidanylisoquinolin-4-yl)carbonyl-4-phenyl-2,7-diazaspiro[4.4]nonan-7-yl]benzamide | | Authors: | Hou, N, Peng, C, Hu, Q. | | Deposit date: | 2021-04-16 | | Release date: | 2022-07-20 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Development of Highly Potent Noncovalent Inhibitors of SARS-CoV-2 3CLpro.
Acs Cent.Sci., 9, 2023
|
|
7EN8
 
 | | Crystal structure of SARS-CoV-2 3CLpro in complex with the non-covalent inhibitor WU-04 | | Descriptor: | 3C-like proteinase, GLYCEROL, ~{N}-[(1~{S},2~{R})-2-[[4-bromanyl-2-(methylcarbamoyl)-6-nitro-phenyl]amino]cyclohexyl]isoquinoline-4-carboxamide | | Authors: | Hou, N, Peng, C, Hu, Q. | | Deposit date: | 2021-04-16 | | Release date: | 2022-07-20 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.83 Å) | | Cite: | Development of Highly Potent Noncovalent Inhibitors of SARS-CoV-2 3CLpro.
Acs Cent.Sci., 9, 2023
|
|
7ENE
 
 | |
7F9Y
 
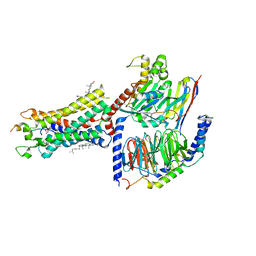 | | ghrelin-bound ghrelin receptor in complex with Gq | | Descriptor: | CHOLESTEROL, Engineered G-alpha-q subunit, Ghrelin-28, ... | | Authors: | Wang, Y, Zhuang, Y, Xu, P, Xu, H.E, Jiang, Y. | | Deposit date: | 2021-07-05 | | Release date: | 2021-08-18 | | Last modified: | 2024-10-30 | | Method: | ELECTRON MICROSCOPY (2.9 Å) | | Cite: | Molecular recognition of an acyl-peptide hormone and activation of ghrelin receptor.
Nat Commun, 12, 2021
|
|
7F9Z
 
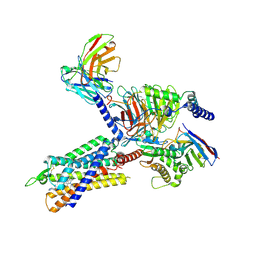 | | GHRP-6-bound ghrelin receptor in complex with Gq | | Descriptor: | 3-PYRIDIN-4-YL-2,4-DIHYDRO-INDENO[1,2-.C.]PYRAZOLE, Engineered G-alpha-q, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, ... | | Authors: | Wang, Y, Zhuang, Y, Xu, P, Xu, H.E, Jiang, Y. | | Deposit date: | 2021-07-05 | | Release date: | 2021-08-18 | | Last modified: | 2025-07-02 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Molecular recognition of an acyl-peptide hormone and activation of ghrelin receptor.
Nat Commun, 12, 2021
|
|
8XOR
 
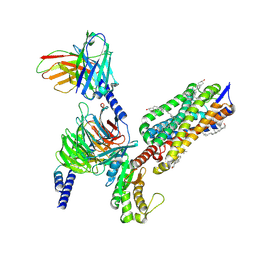 | | Cryo-EM structure of the tethered agonist-bound human PAR1-Gq complex | | Descriptor: | CHOLESTEROL, G subunit q (Gi1-Gq chimeric), Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, ... | | Authors: | Guo, J, Zhang, Y. | | Deposit date: | 2024-01-02 | | Release date: | 2024-09-18 | | Last modified: | 2024-11-20 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | Structural basis of tethered agonism and G protein coupling of protease-activated receptors.
Cell Res., 34, 2024
|
|
8XOS
 
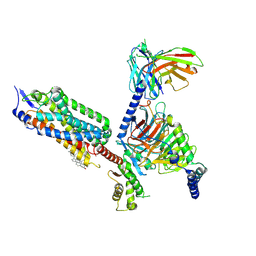 | | Cryo-EM structure of the tethered agonist-bound human PAR1-Gi complex | | Descriptor: | CHOLESTEROL, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, ... | | Authors: | Guo, J, Zhang, Y. | | Deposit date: | 2024-01-02 | | Release date: | 2024-09-18 | | Last modified: | 2025-06-18 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Structural basis of tethered agonism and G protein coupling of protease-activated receptors.
Cell Res., 34, 2024
|
|
7XMW
 
 | | Crystal structure of anti-CRISPR protein AcrVIA2 | | Descriptor: | 2-{2-[2-(2-{2-[2-(2-ETHOXY-ETHOXY)-ETHOXY]-ETHOXY}-ETHOXY)-ETHOXY]-ETHOXY}-ETHANOL, AcrVIA2, SELENIUM ATOM, ... | | Authors: | Yan, X, Li, X, Song, G. | | Deposit date: | 2022-04-27 | | Release date: | 2023-05-31 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.59 Å) | | Cite: | Structure of AcrVIA2 and its binding mechanism to CRISPR-Cas13a.
Biochem.Biophys.Res.Commun., 612, 2022
|
|
7V9P
 
 | |
7V9Q
 
 | |
7V9O
 
 | |
