6EP5
 
 | |
6EP2
 
 | |
6ER8
 
 | |
1FS6
 
 | | GLUCOSAMINE-6-PHOSPHATE DEAMINASE FROM E.COLI, T CONFORMER, AT 2.2A RESOLUTION | | Descriptor: | GLUCOSAMINE-6-PHOSPHATE DEAMINASE | | Authors: | Rudino-Pinera, E, Morales-Arrieta, S, Rojas-Trejo, S.P, Horjales, E. | | Deposit date: | 2000-09-08 | | Release date: | 2002-01-04 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structural flexibility, an essential component of the allosteric activation in Escherichia coli glucosamine-6-phosphate deaminase.
Acta Crystallogr.,Sect.D, 58, 2002
|
|
1FS5
 
 | | A DISCOVERY OF THREE ALTERNATE CONFORMATIONS IN THE ACTIVE SITE OF GLUCOSAMINE-6-PHOSPHATE ISOMERASE | | Descriptor: | 2-acetamido-2-deoxy-6-O-phosphono-alpha-D-glucopyranose, GLUCOSAMINE-6-PHOSPHATE DEAMINASE, L(+)-TARTARIC ACID | | Authors: | Rudino-Pinera, E, Morales-Arrieta, S, Rojas-Trejo, S.P, Horjales, E. | | Deposit date: | 2000-09-08 | | Release date: | 2002-01-04 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.73 Å) | | Cite: | Structural flexibility, an essential component of the allosteric activation in Escherichia coli glucosamine-6-phosphate deaminase.
Acta Crystallogr.,Sect.D, 58, 2002
|
|
1FQO
 
 | | GLUCOSAMINE 6-PHOSPHATE DEAMINASE COMPLEXED WITH THE SUBSTRATE OF THE REVERSE REACTION FRUCTOSE 6-PHOSPHATE (OPEN FORM) | | Descriptor: | FRUCTOSE -6-PHOSPHATE, GLUCOSAMINE-6-PHOSPHATE DEAMINASE | | Authors: | Rudino-Pinera, E, Morales-Arrieta, S, Rojas-Trejo, S.P, Horjales, E. | | Deposit date: | 2000-09-06 | | Release date: | 2002-01-04 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structural flexibility, an essential component of the allosteric activation in Escherichia coli glucosamine-6-phosphate deaminase.
Acta Crystallogr.,Sect.D, 58, 2002
|
|
1FRZ
 
 | | GLUCOSAMINE-6-PHOSPHATE DEAMINASE FROM E.COLI, R CONFORMER. COMPLEXED WITH THE ALLOSTERIC ACTIVATOR N-ACETYL-GLUCOSAMINE-6-PHOSPHATE AT 2.2 A RESOLUTION | | Descriptor: | 2-acetamido-2-deoxy-6-O-phosphono-alpha-D-glucopyranose, GLUCOSAMINE-6-PHOSPHATE DEAMINASE | | Authors: | Rudino-Pinera, E, Morales-Arrieta, S, Rojas-Trejo, S.P, Horjales, E. | | Deposit date: | 2000-09-07 | | Release date: | 2002-01-04 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structural flexibility, an essential component of the allosteric activation in Escherichia coli glucosamine-6-phosphate deaminase.
Acta Crystallogr.,Sect.D, 58, 2002
|
|
1FSF
 
 | | GLUCOSAMINE-6-PHOSPHATE DEAMINASE FROM E.COLI, T CONFORMER, AT 1.9A RESOLUTION | | Descriptor: | GLUCOSAMINE-6-PHOSPHATE DEAMINASE | | Authors: | Rudino-Pinera, E, Morales-Arrieta, S, Rojas-Trejo, S.P, Horjales, E. | | Deposit date: | 2000-09-08 | | Release date: | 2002-01-04 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structural flexibility, an essential component of the allosteric activation in Escherichia coli glucosamine-6-phosphate deaminase.
Acta Crystallogr.,Sect.D, 58, 2002
|
|
5NV5
 
 | | Enterococcus faecalis FIC protein | | Descriptor: | Fic family protein | | Authors: | Veyron, S, Cherfils, J. | | Deposit date: | 2017-05-03 | | Release date: | 2018-05-30 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | A Ca2+-regulated deAMPylation switch in human and bacterial FIC proteins.
Nat Commun, 10, 2019
|
|
5NWF
 
 | |
1JLH
 
 | | Human Glucose-6-phosphate Isomerase | | Descriptor: | phosphoglucose isomerase | | Authors: | Cordeiro, A.T. | | Deposit date: | 2001-07-16 | | Release date: | 2003-02-11 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Crystal structure of human phosphoglucose isomerase and analysis of the initial catalytic steps
BIOCHIM.BIOPHYS.ACTA, 1645, 2003
|
|
1ORE
 
 | | Human Adenine Phosphoribosyltransferase | | Descriptor: | ADENOSINE MONOPHOSPHATE, Adenine phosphoribosyltransferase, CHLORIDE ION | | Authors: | Thiemann, O.H, Silva, M, Oliva, G, Silva, C.H.T.P, Iulek, J. | | Deposit date: | 2003-03-12 | | Release date: | 2004-03-30 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Three-dimensional structure of human adenine phosphoribosyltransferase and its relation to DHA-urolithiasis.
Biochemistry, 43, 2004
|
|
1NE7
 
 | | HUMAN GLUCOSAMINE-6-PHOSPHATE DEAMINASE ISOMERASE AT 1.75 A RESOLUTION COMPLEXED WITH N-ACETYL-GLUCOSAMINE-6-PHOSPHATE AND 2-DEOXY-2-AMINO-GLUCITOL-6-PHOSPHATE | | Descriptor: | 2-DEOXY-2-AMINO GLUCITOL-6-PHOSPHATE, 2-acetamido-2-deoxy-6-O-phosphono-alpha-D-glucopyranose, Glucosamine-6-phosphate isomerase, ... | | Authors: | Arreola, R, Valderrama, B, Morante, M.L, Horjales, E. | | Deposit date: | 2002-12-10 | | Release date: | 2003-09-23 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Two mammalian glucosamine-6-phosphate deaminases: a structural and genetic study.
Febs Lett., 551, 2003
|
|
1GQ0
 
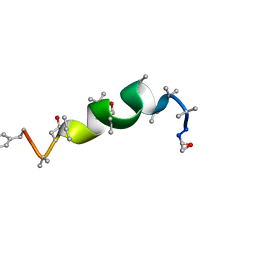 | | Solution structure of Antiamoebin I, a membrane channel-forming polypeptide; NMR, 20 structures | | Descriptor: | ANTIAMOEBIN I | | Authors: | Galbraith, T.P, Harris, R, Driscoll, P.C, Wallace, B.A. | | Deposit date: | 2001-11-16 | | Release date: | 2003-01-24 | | Last modified: | 2017-12-20 | | Method: | SOLUTION NMR | | Cite: | Solution NMR studies of antiamoebin, a membrane channel-forming polypeptide.
Biophys. J., 84, 2003
|
|
1LGN
 
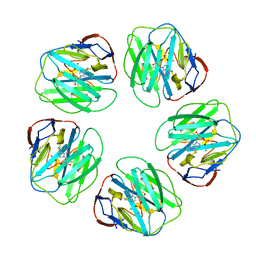 | |
7GH9
 
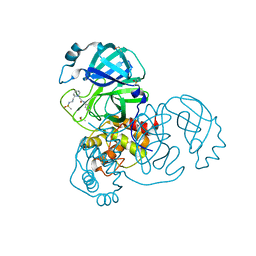 | | Group deposition SARS-CoV-2 main protease in complex with inhibitors from the COVID Moonshot -- Crystal Structure of SARS-CoV-2 main protease in complex with BEN-DND-362d364a-10 (Mpro-x2971) | | Descriptor: | 2-[(2S)-2-{2-[(methanesulfonyl)amino]ethyl}piperidin-1-yl]-N-(pyridin-3-yl)acetamide, 3C-like proteinase, DIMETHYL SULFOXIDE | | Authors: | Fearon, D, Aimon, A, Aschenbrenner, J.C, Balcomb, B.H, Bertram, F.K.R, Brandao-Neto, J, Dias, A, Douangamath, A, Dunnett, L, Godoy, A.S, Gorrie-Stone, T.J, Koekemoer, L, Krojer, T, Lithgo, R.M, Lukacik, P, Marples, P.G, Mikolajek, H, Nelson, E, Owen, C.D, Powell, A.J, Rangel, V.L, Skyner, R, Strain-Damerell, C.M, Thompson, W, Tomlinson, C.W.E, Wild, C, Walsh, M.A, von Delft, F. | | Deposit date: | 2023-08-11 | | Release date: | 2023-11-08 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (1.57 Å) | | Cite: | Open science discovery of potent noncovalent SARS-CoV-2 main protease inhibitors.
Science, 382, 2023
|
|
7GHG
 
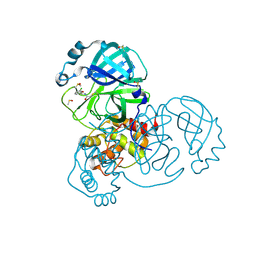 | | Group deposition SARS-CoV-2 main protease in complex with inhibitors from the COVID Moonshot -- Crystal Structure of SARS-CoV-2 main protease in complex with DAV-CRI-3edb475e-4 (Mpro-x3324) | | Descriptor: | 3C-like proteinase, DIMETHYL SULFOXIDE, N-[(1R)-1-(3-chlorophenyl)-2-hydroxyethyl]acetamide | | Authors: | Fearon, D, Aimon, A, Aschenbrenner, J.C, Balcomb, B.H, Bertram, F.K.R, Brandao-Neto, J, Dias, A, Douangamath, A, Dunnett, L, Godoy, A.S, Gorrie-Stone, T.J, Koekemoer, L, Krojer, T, Lithgo, R.M, Lukacik, P, Marples, P.G, Mikolajek, H, Nelson, E, Owen, C.D, Powell, A.J, Rangel, V.L, Skyner, R, Strain-Damerell, C.M, Thompson, W, Tomlinson, C.W.E, Wild, C, Walsh, M.A, von Delft, F. | | Deposit date: | 2023-08-11 | | Release date: | 2023-11-08 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | Open science discovery of potent noncovalent SARS-CoV-2 main protease inhibitors.
Science, 382, 2023
|
|
7GD7
 
 | | Group deposition SARS-CoV-2 main protease in complex with inhibitors from the COVID Moonshot -- Crystal Structure of SARS-CoV-2 main protease in complex with ADA-UNI-f8e79267-2 (Mpro-x10889) | | Descriptor: | (2R)-4-[(methanesulfonyl)amino]-2-phenyl-N-(pyridin-3-yl)butanamide, 3C-like proteinase, DIMETHYL SULFOXIDE | | Authors: | Fearon, D, Aimon, A, Aschenbrenner, J.C, Balcomb, B.H, Bertram, F.K.R, Brandao-Neto, J, Dias, A, Douangamath, A, Dunnett, L, Godoy, A.S, Gorrie-Stone, T.J, Koekemoer, L, Krojer, T, Lithgo, R.M, Lukacik, P, Marples, P.G, Mikolajek, H, Nelson, E, Owen, C.D, Powell, A.J, Rangel, V.L, Skyner, R, Strain-Damerell, C.M, Thompson, W, Tomlinson, C.W.E, Wild, C, Walsh, M.A, von Delft, F. | | Deposit date: | 2023-08-11 | | Release date: | 2023-11-08 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (1.509 Å) | | Cite: | Open science discovery of potent noncovalent SARS-CoV-2 main protease inhibitors.
Science, 382, 2023
|
|
7GHH
 
 | | Group deposition SARS-CoV-2 main protease in complex with inhibitors from the COVID Moonshot -- Crystal Structure of SARS-CoV-2 main protease in complex with TOB-UNK-c2aba166-1 (Mpro-x3325) | | Descriptor: | 1-[4-(prop-2-yn-1-yl)piperazin-1-yl]ethan-1-one, 3C-like proteinase, DIMETHYL SULFOXIDE | | Authors: | Fearon, D, Aimon, A, Aschenbrenner, J.C, Balcomb, B.H, Bertram, F.K.R, Brandao-Neto, J, Dias, A, Douangamath, A, Dunnett, L, Godoy, A.S, Gorrie-Stone, T.J, Koekemoer, L, Krojer, T, Lithgo, R.M, Lukacik, P, Marples, P.G, Mikolajek, H, Nelson, E, Owen, C.D, Powell, A.J, Rangel, V.L, Skyner, R, Strain-Damerell, C.M, Thompson, W, Tomlinson, C.W.E, Wild, C, Walsh, M.A, von Delft, F. | | Deposit date: | 2023-08-11 | | Release date: | 2023-11-08 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (1.56 Å) | | Cite: | Open science discovery of potent noncovalent SARS-CoV-2 main protease inhibitors.
Science, 382, 2023
|
|
7GDH
 
 | | Group deposition SARS-CoV-2 main protease in complex with inhibitors from the COVID Moonshot -- Crystal Structure of SARS-CoV-2 main protease in complex with LOR-NEU-c8f11034-6 (Mpro-x11001) | | Descriptor: | (3S)-3-hydroxy-2-oxo-2,3-dihydro-1H-indole-5-sulfonamide, 3C-like proteinase, DIMETHYL SULFOXIDE | | Authors: | Fearon, D, Aimon, A, Aschenbrenner, J.C, Balcomb, B.H, Bertram, F.K.R, Brandao-Neto, J, Dias, A, Douangamath, A, Dunnett, L, Godoy, A.S, Gorrie-Stone, T.J, Koekemoer, L, Krojer, T, Lithgo, R.M, Lukacik, P, Marples, P.G, Mikolajek, H, Nelson, E, Owen, C.D, Powell, A.J, Rangel, V.L, Skyner, R, Strain-Damerell, C.M, Thompson, W, Tomlinson, C.W.E, Wild, C, Walsh, M.A, von Delft, F. | | Deposit date: | 2023-08-11 | | Release date: | 2023-11-08 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (1.891 Å) | | Cite: | Open science discovery of potent noncovalent SARS-CoV-2 main protease inhibitors.
Science, 382, 2023
|
|
7GB2
 
 | | Group deposition SARS-CoV-2 main protease in complex with inhibitors from the COVID Moonshot -- Crystal Structure of SARS-CoV-2 main protease in complex with MED-COV-4280ac29-25 (Mpro-x10155) | | Descriptor: | 1-{4-[(2-benzyl-1,3-thiazol-5-yl)methyl]piperazin-1-yl}ethan-1-one, 3C-like proteinase, DIMETHYL SULFOXIDE | | Authors: | Fearon, D, Aimon, A, Aschenbrenner, J.C, Balcomb, B.H, Bertram, F.K.R, Brandao-Neto, J, Dias, A, Douangamath, A, Dunnett, L, Godoy, A.S, Gorrie-Stone, T.J, Koekemoer, L, Krojer, T, Lithgo, R.M, Lukacik, P, Marples, P.G, Mikolajek, H, Nelson, E, Owen, C.D, Powell, A.J, Rangel, V.L, Skyner, R, Strain-Damerell, C.M, Thompson, W, Tomlinson, C.W.E, Wild, C, Walsh, M.A, von Delft, F. | | Deposit date: | 2023-08-11 | | Release date: | 2023-11-08 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Open science discovery of potent noncovalent SARS-CoV-2 main protease inhibitors.
Science, 382, 2023
|
|
7GDN
 
 | | Group deposition SARS-CoV-2 main protease in complex with inhibitors from the COVID Moonshot -- Crystal Structure of SARS-CoV-2 main protease in complex with EDJ-MED-0e996074-1 (Mpro-x11159) | | Descriptor: | (4R)-N-(4-cyclopropyl-4H-1,2,4-triazol-3-yl)-3,4-dihydro-2H-1-benzopyran-4-carboxamide, 3C-like proteinase, DIMETHYL SULFOXIDE | | Authors: | Fearon, D, Aimon, A, Aschenbrenner, J.C, Balcomb, B.H, Bertram, F.K.R, Brandao-Neto, J, Dias, A, Douangamath, A, Dunnett, L, Godoy, A.S, Gorrie-Stone, T.J, Koekemoer, L, Krojer, T, Lithgo, R.M, Lukacik, P, Marples, P.G, Mikolajek, H, Nelson, E, Owen, C.D, Powell, A.J, Rangel, V.L, Skyner, R, Strain-Damerell, C.M, Thompson, W, Tomlinson, C.W.E, Wild, C, Walsh, M.A, von Delft, F. | | Deposit date: | 2023-08-11 | | Release date: | 2023-11-08 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (1.351 Å) | | Cite: | Open science discovery of potent noncovalent SARS-CoV-2 main protease inhibitors.
Science, 382, 2023
|
|
7GBI
 
 | | Group deposition SARS-CoV-2 main protease in complex with inhibitors from the COVID Moonshot -- Crystal Structure of SARS-CoV-2 main protease in complex with JAG-UCB-a3ef7265-3 (Mpro-x10338) | | Descriptor: | (3S)-5-chloro-N-[4-(hydroxymethyl)pyridin-3-yl]-2,3-dihydro-1-benzofuran-3-carboxamide, 3C-like proteinase, DIMETHYL SULFOXIDE | | Authors: | Fearon, D, Aimon, A, Aschenbrenner, J.C, Balcomb, B.H, Bertram, F.K.R, Brandao-Neto, J, Dias, A, Douangamath, A, Dunnett, L, Godoy, A.S, Gorrie-Stone, T.J, Koekemoer, L, Krojer, T, Lithgo, R.M, Lukacik, P, Marples, P.G, Mikolajek, H, Nelson, E, Owen, C.D, Powell, A.J, Rangel, V.L, Skyner, R, Strain-Damerell, C.M, Thompson, W, Tomlinson, C.W.E, Wild, C, Walsh, M.A, von Delft, F. | | Deposit date: | 2023-08-11 | | Release date: | 2023-11-08 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (1.291 Å) | | Cite: | Open science discovery of potent noncovalent SARS-CoV-2 main protease inhibitors.
Science, 382, 2023
|
|
7GDY
 
 | | Group deposition SARS-CoV-2 main protease in complex with inhibitors from the COVID Moonshot -- Crystal Structure of SARS-CoV-2 main protease in complex with LOR-NOR-30067bb9-6 (Mpro-x11258) | | Descriptor: | (3S)-5-bromo-1-[(2-ethoxyphenyl)methyl]-3-hydroxy-1,3-dihydro-2H-indol-2-one, 3C-like proteinase, DIMETHYL SULFOXIDE | | Authors: | Fearon, D, Aimon, A, Aschenbrenner, J.C, Balcomb, B.H, Bertram, F.K.R, Brandao-Neto, J, Dias, A, Douangamath, A, Dunnett, L, Godoy, A.S, Gorrie-Stone, T.J, Koekemoer, L, Krojer, T, Lithgo, R.M, Lukacik, P, Marples, P.G, Mikolajek, H, Nelson, E, Owen, C.D, Powell, A.J, Rangel, V.L, Skyner, R, Strain-Damerell, C.M, Thompson, W, Tomlinson, C.W.E, Wild, C, Walsh, M.A, von Delft, F. | | Deposit date: | 2023-08-11 | | Release date: | 2023-11-08 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (1.611 Å) | | Cite: | Open science discovery of potent noncovalent SARS-CoV-2 main protease inhibitors.
Science, 382, 2023
|
|
7GBZ
 
 | | Group deposition SARS-CoV-2 main protease in complex with inhibitors from the COVID Moonshot -- Crystal Structure of SARS-CoV-2 main protease in complex with MAT-POS-590ac91e-11 (Mpro-x10474) | | Descriptor: | (3S)-3,4-dimethyl-N-(4-methylpyridin-3-yl)pentanamide, 3C-like proteinase, DIMETHYL SULFOXIDE | | Authors: | Fearon, D, Aimon, A, Aschenbrenner, J.C, Balcomb, B.H, Bertram, F.K.R, Brandao-Neto, J, Dias, A, Douangamath, A, Dunnett, L, Godoy, A.S, Gorrie-Stone, T.J, Koekemoer, L, Krojer, T, Lithgo, R.M, Lukacik, P, Marples, P.G, Mikolajek, H, Nelson, E, Owen, C.D, Powell, A.J, Rangel, V.L, Skyner, R, Strain-Damerell, C.M, Thompson, W, Tomlinson, C.W.E, Wild, C, Walsh, M.A, von Delft, F. | | Deposit date: | 2023-08-11 | | Release date: | 2023-11-08 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (1.529 Å) | | Cite: | Open science discovery of potent noncovalent SARS-CoV-2 main protease inhibitors.
Science, 382, 2023
|
|
