5GXQ
 
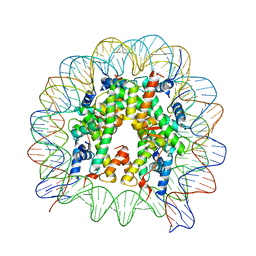 | | The crystal structure of the nucleosome containing H3.6 | | Descriptor: | DNA (146-MER), Histone H2A type 1-B/E, Histone H2B type 1-J, ... | | Authors: | Taguchi, H, Xie, Y, Horikoshi, N, Kurumizaka, H. | | Deposit date: | 2016-09-19 | | Release date: | 2017-04-19 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.85 Å) | | Cite: | Crystal Structure and Characterization of Novel Human Histone H3 Variants, H3.6, H3.7, and H3.8
Biochemistry, 56, 2017
|
|
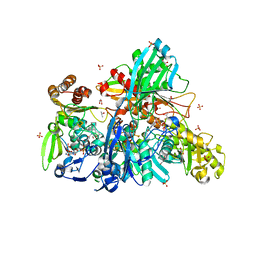
5Y6Q
 
 | |
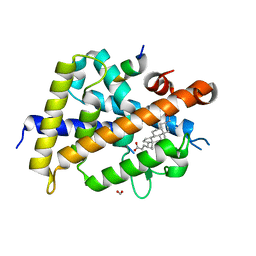
7C7V
 
 | | Vitamin D3 receptor/lithochoric acid derivative complex | | Descriptor: | (4R)-4-[(3R,5R,8R,9S,10S,13R,14S,17R)-10,13-dimethyl-3-(2-methyl-2-oxidanyl-propyl)-2,3,4,5,6,7,8,9,11,12,14,15,16,17-tetradecahydro-1H-cyclopenta[a]phenanthren-17-yl]pentanoic acid, FORMIC ACID, Mediator of RNA polymerase II transcription subunit 1, ... | | Authors: | Masuno, H, Numoto, N, Kagechika, H, Ito, N. | | Deposit date: | 2020-05-26 | | Release date: | 2021-01-20 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Lithocholic Acid Derivatives as Potent Vitamin D Receptor Agonists.
J.Med.Chem., 64, 2021
|
|
7C7W
 
 | | Vitamin D3 receptor/lithochoric acid derivative complex | | Descriptor: | (4R)-4-[(3S,5R,8R,9S,10S,13R,14S,17R)-10,13-dimethyl-3-(2-methyl-2-oxidanyl-propyl)-2,3,4,5,6,7,8,9,11,12,14,15,16,17-tetradecahydro-1H-cyclopenta[a]phenanthren-17-yl]pentanoic acid, FORMIC ACID, Mediator of RNA polymerase II transcription subunit 1, ... | | Authors: | Masuno, H, Numoto, N, Kagechika, H, Ito, N. | | Deposit date: | 2020-05-26 | | Release date: | 2021-01-20 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Lithocholic Acid Derivatives as Potent Vitamin D Receptor Agonists.
J.Med.Chem., 64, 2021
|
|
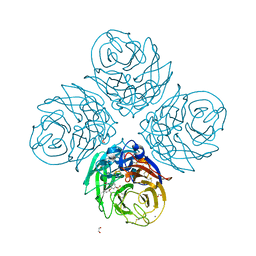
6G01
 
 | |
6G02
 
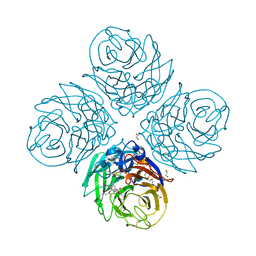 | | Complex of neuraminidase from H1N1 influenza virus with tamiphosphor omega-azidohexyl ester | | Descriptor: | 1,2-ETHANEDIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose, 6-[[(3~{R},4~{R},5~{S})-4-acetamido-5-azanyl-3-pentan-3-yloxy-cyclohexen-1-yl]-oxidanyl-phosphoryl]oxyhexylimino-azanylidene-azanium, ... | | Authors: | Pachl, P, Pokorna, J. | | Deposit date: | 2018-03-15 | | Release date: | 2019-01-23 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (1.84 Å) | | Cite: | DNA-linked inhibitor antibody assay (DIANA) as a new method for screening influenza neuraminidase inhibitors.
Biochem. J., 475, 2018
|
|
5B1M
 
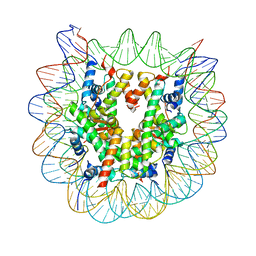 | | The mouse nucleosome structure containing H3.1 | | Descriptor: | DNA (146-MER), Histone H2A type 1, Histone H2B type 3-A, ... | | Authors: | Urahama, T, Machida, S, Horikoshi, N, Osakabe, A, Tachiwana, H, Taguchi, H, Kurumizaka, H. | | Deposit date: | 2015-12-08 | | Release date: | 2017-02-15 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.34 Å) | | Cite: | Testis-Specific Histone Variant H3t Gene Is Essential for Entry into Spermatogenesis
Cell Rep, 18, 2017
|
|
1CYC
 
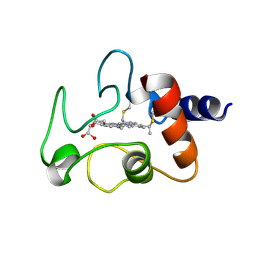 | | THE CRYSTAL STRUCTURE OF BONITO (KATSUO) FERROCYTOCHROME C AT 2.3 ANGSTROMS RESOLUTION. II. STRUCTURE AND FUNCTION | | Descriptor: | FERROCYTOCHROME C, HEME C | | Authors: | Tanaka, N, Yamane, T, Tsukihara, T, Ashida, T, Kakudo, M. | | Deposit date: | 1976-08-01 | | Release date: | 1976-10-06 | | Last modified: | 2021-03-03 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | The crystal structure of bonito (katsuo) ferrocytochrome c at 2.3 A resolution. II. Structure and function.
J.Biochem.(Tokyo), 77, 1975
|
|
5YC5
 
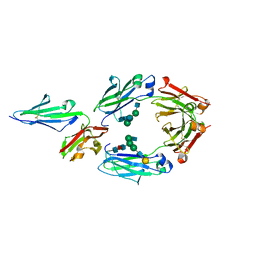 | | Crystal structure of human IgG-Fc in complex with aglycan and optimized Fc gamma receptor IIIa | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-2)-alpha-D-mannopyranose-(1-3)-[2-acetamido-2-deoxy-beta-D-glucopyranose-(1-2)-alpha-D-mannopyranose-(1-6)]beta-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, CHLORIDE ION, Immunoglobulin gamma-1 heavy chain, ... | | Authors: | Caaveiro, J.M.M, Tamura, H, Tsumoto, K, Kiyoshi, M. | | Deposit date: | 2017-09-06 | | Release date: | 2018-03-21 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.71 Å) | | Cite: | Assessing the Heterogeneity of the Fc-Glycan of a Therapeutic Antibody Using an engineered Fc gamma Receptor IIIa-Immobilized Column.
Sci Rep, 8, 2018
|
|
6P64
 
 | |
8D0Z
 
 | | S728-1157 IgG in complex with SARS-CoV-2-6P-Mut7 Spike protein (focused refinement) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, S728-1157 Fab heavy chain variable region, S728-1157 Fab light chain variable region, ... | | Authors: | Ozorowski, G, Torres, J.L, Ward, A.B. | | Deposit date: | 2022-05-26 | | Release date: | 2023-03-22 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (3.7 Å) | | Cite: | Site of vulnerability on SARS-CoV-2 spike induces broadly protective antibody against antigenically distinct Omicron subvariants.
J.Clin.Invest., 133, 2023
|
|
8R7A
 
 | |
6SYI
 
 | |
6YEM
 
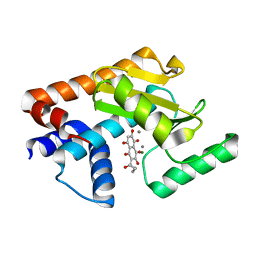 | | H1N1 2009 PA Endonuclease in complex with Quambalarine B | | Descriptor: | 3,5,6,8-tetrakis(oxidanyl)-2-pentanoyl-naphthalene-1,4-dione, CHLORIDE ION, MAGNESIUM ION, ... | | Authors: | Radilova, K, Brynda, J. | | Deposit date: | 2020-03-25 | | Release date: | 2020-09-09 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Unraveling the anti-influenza effect of flavonoids: Experimental validation of luteolin and its congeners as potent influenza endonuclease inhibitors.
Eur.J.Med.Chem., 208, 2020
|
|
1EJ2
 
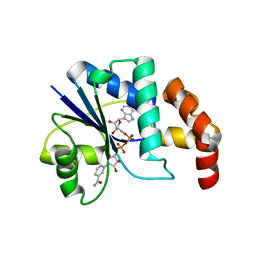 | | Crystal structure of methanobacterium thermoautotrophicum nicotinamide mononucleotide adenylyltransferase with bound NAD+ | | Descriptor: | NICOTINAMIDE MONONUCLEOTIDE ADENYLYLTRANSFERASE, NICOTINAMIDE-ADENINE-DINUCLEOTIDE, SODIUM ION, ... | | Authors: | Saridakis, V, Christendat, D, Kimber, M.S, Edwards, A.M, Pai, E.F, Midwest Center for Structural Genomics (MCSG), Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2000-02-29 | | Release date: | 2001-03-14 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Insights into ligand binding and catalysis of a central step in NAD+ synthesis: structures of Methanobacterium thermoautotrophicum NMN adenylyltransferase complexes.
J.Biol.Chem., 276, 2001
|
|
8R7D
 
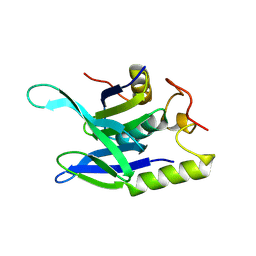 | |
6YA5
 
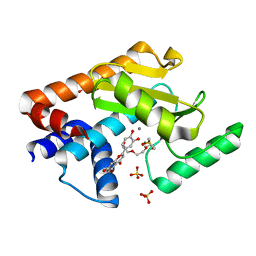 | | 2009 H1N1 PA Endonuclease in complex with LU2 | | Descriptor: | 2-(3,4-dihydroxyphenyl)-5,7-dihydroxy-4H-chromen-4-one, DI(HYDROXYETHYL)ETHER, DIMETHYL SULFOXIDE, ... | | Authors: | Radilova, K, Brynda, J. | | Deposit date: | 2020-03-11 | | Release date: | 2020-09-09 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Unraveling the anti-influenza effect of flavonoids: Experimental validation of luteolin and its congeners as potent influenza endonuclease inhibitors.
Eur.J.Med.Chem., 208, 2020
|
|
7ZPY
 
 | |
5YO8
 
 | |
7EM8
 
 | | Crystal structure of the PI5P4Kbeta T201M-2a-ATP complex | | Descriptor: | Phosphatidylinositol 5-phosphate 4-kinase type-2 beta, [(2~{R},3~{S},4~{R},5~{R})-5-[2,6-bis(azanyl)purin-9-yl]-3,4-bis(oxidanyl)oxolan-2-yl]methyl phosphono hydrogen phosphate, [[(2~{R},3~{S},4~{R},5~{R})-5-[2,6-bis(azanyl)purin-9-yl]-3,4-bis(oxidanyl)oxolan-2-yl]methoxy-oxidanyl-phosphoryl] phosphono hydrogen phosphate | | Authors: | Senda, M, Senda, T. | | Deposit date: | 2021-04-13 | | Release date: | 2022-03-30 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (3.05 Å) | | Cite: | The GTP responsiveness of PI5P4K beta evolved from a compromised trade-off between activity and specificity.
Structure, 30, 2022
|
|
7EM2
 
 | | Crystal structure of the PI5P4Kbeta-XTP complex | | Descriptor: | Phosphatidylinositol 5-phosphate 4-kinase type-2 beta, [(2~{R},3~{S},4~{R},5~{R})-5-[2,6-bis(oxidanylidene)-3~{H}-purin-9-yl]-3,4-bis(oxidanyl)oxolan-2-yl]methyl phosphono hydrogen phosphate, [[(2~{R},3~{S},4~{R},5~{R})-5-[2,6-bis(oxidanylidene)-3~{H}-purin-9-yl]-3,4-bis(oxidanyl)oxolan-2-yl]methoxy-oxidanyl-phosphoryl] phosphono hydrogen phosphate | | Authors: | Senda, M, Senda, T. | | Deposit date: | 2021-04-13 | | Release date: | 2022-03-30 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | The GTP responsiveness of PI5P4K beta evolved from a compromised trade-off between activity and specificity.
Structure, 30, 2022
|
|
7EM5
 
 | | Crystal structure of the PI5P4Kbeta F205L-XTP complex | | Descriptor: | Phosphatidylinositol 5-phosphate 4-kinase type-2 beta, [(2~{R},3~{S},4~{R},5~{R})-5-[2,6-bis(oxidanylidene)-3~{H}-purin-9-yl]-3,4-bis(oxidanyl)oxolan-2-yl]methyl phosphono hydrogen phosphate, [[(2~{R},3~{S},4~{R},5~{R})-5-[2,6-bis(oxidanylidene)-3~{H}-purin-9-yl]-3,4-bis(oxidanyl)oxolan-2-yl]methoxy-oxidanyl-phosphoryl] phosphono hydrogen phosphate | | Authors: | Senda, M, Senda, T. | | Deposit date: | 2021-04-13 | | Release date: | 2022-03-30 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | The GTP responsiveness of PI5P4K beta evolved from a compromised trade-off between activity and specificity.
Structure, 30, 2022
|
|
7EM1
 
 | | Crystal structure of the PI5P4Kbeta-ITP complex | | Descriptor: | INOSINE-5'-DIPHOSPHATE, Phosphatidylinositol 5-phosphate 4-kinase type-2 beta, [[(2~{R},3~{S},4~{R},5~{R})-3,4-bis(oxidanyl)-5-(6-oxidanylidene-1~{H}-purin-9-yl)oxolan-2-yl]methoxy-oxidanyl-phosphoryl] phosphono hydrogen phosphate | | Authors: | Senda, M, Senda, T. | | Deposit date: | 2021-04-13 | | Release date: | 2022-03-30 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.65 Å) | | Cite: | The GTP responsiveness of PI5P4K beta evolved from a compromised trade-off between activity and specificity.
Structure, 30, 2022
|
|
7EM6
 
 | | Crystal structure of the PI5P4Kbeta N203D-ITP complex | | Descriptor: | INOSINE-5'-DIPHOSPHATE, Phosphatidylinositol 5-phosphate 4-kinase type-2 beta, [[(2~{R},3~{S},4~{R},5~{R})-3,4-bis(oxidanyl)-5-(6-oxidanylidene-1~{H}-purin-9-yl)oxolan-2-yl]methoxy-oxidanyl-phosphoryl] phosphono hydrogen phosphate | | Authors: | Senda, M, Senda, T. | | Deposit date: | 2021-04-13 | | Release date: | 2022-03-30 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.95 Å) | | Cite: | The GTP responsiveness of PI5P4K beta evolved from a compromised trade-off between activity and specificity.
Structure, 30, 2022
|
|
7EM7
 
 | | Crystal structure of the PI5P4Kbeta N203D-XTP complex | | Descriptor: | Phosphatidylinositol 5-phosphate 4-kinase type-2 beta, [(2~{R},3~{S},4~{R},5~{R})-5-[2,6-bis(oxidanylidene)-3~{H}-purin-9-yl]-3,4-bis(oxidanyl)oxolan-2-yl]methyl phosphono hydrogen phosphate, [[(2~{R},3~{S},4~{R},5~{R})-5-[2,6-bis(oxidanylidene)-3~{H}-purin-9-yl]-3,4-bis(oxidanyl)oxolan-2-yl]methoxy-oxidanyl-phosphoryl] phosphono hydrogen phosphate | | Authors: | Senda, M, Senda, T. | | Deposit date: | 2021-04-13 | | Release date: | 2022-03-30 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (3.45 Å) | | Cite: | The GTP responsiveness of PI5P4K beta evolved from a compromised trade-off between activity and specificity.
Structure, 30, 2022
|
|
