5FDP
 
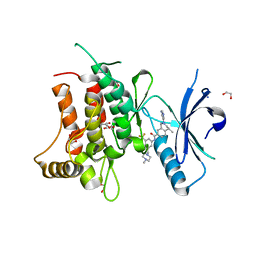 | | Structure of DDR1 receptor tyrosine kinase in complex with D2099 inhibitor at 2.25 Angstroms resolution. | | Descriptor: | (4~{S})-4-methyl-~{N}-[3-[(4-methylpiperazin-1-yl)methyl]-5-(trifluoromethyl)phenyl]-2-pyrimidin-5-yl-3,4-dihydro-1~{H}-isoquinoline-7-carboxamide, 1,2-ETHANEDIOL, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Bartual, S.G, Pinkas, D.M, Wang, Z, Ding, K, Mahajan, P, Kupinska, K, Mukhopadhyay, S, Strain-Damerell, C, Borkowska, O, Talon, R, Kopec, J, Williams, E, Tallant, C, Chaikuad, A, Sorell, F, Newman, J, Burgess-Brown, N, Arrowsmith, C.H, von Delft, F, Edwards, A.M, Bountra, C, Bullock, A, Structural Genomics Consortium (SGC) | | Deposit date: | 2015-12-16 | | Release date: | 2016-06-08 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Structure-Based Design of Tetrahydroisoquinoline-7-carboxamides as Selective Discoidin Domain Receptor 1 (DDR1) Inhibitors.
J.Med.Chem., 59, 2016
|
|
1CEG
 
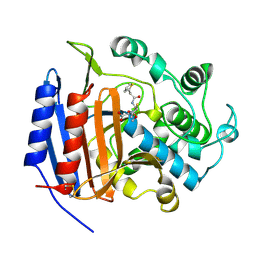 | | CEPHALOTHIN COMPLEXED WITH DD-PEPTIDASE | | Descriptor: | CEPHALOTHIN GROUP, D-ALANYL-D-ALANINE CARBOXYPEPTIDASE TRANSPEPTIDASE | | Authors: | Knox, J.R, Kuzin, A.P. | | Deposit date: | 1995-01-12 | | Release date: | 1996-10-14 | | Last modified: | 2024-06-05 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Binding of cephalothin and cefotaxime to D-ala-D-ala-peptidase reveals a functional basis of a natural mutation in a low-affinity penicillin-binding protein and in extended-spectrum beta-lactamases.
Biochemistry, 34, 1995
|
|
1CEF
 
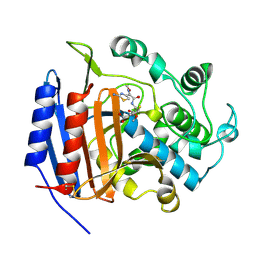 | | CEFOTAXIME COMPLEXED WITH THE STREPTOMYCES R61 DD-PEPTIDASE | | Descriptor: | CEFOTAXIME, C3' cleaved, open, ... | | Authors: | Knox, J.R, Kuzin, A.P. | | Deposit date: | 1995-01-12 | | Release date: | 1996-10-14 | | Last modified: | 2024-06-19 | | Method: | X-RAY DIFFRACTION (2.04 Å) | | Cite: | Binding of cephalothin and cefotaxime to D-ala-D-ala-peptidase reveals a functional basis of a natural mutation in a low-affinity penicillin-binding protein and in extended-spectrum beta-lactamases.
Biochemistry, 34, 1995
|
|
7MKM
 
 | |
7MKL
 
 | |
4L04
 
 | |
4KZO
 
 | |
4L06
 
 | |
3O50
 
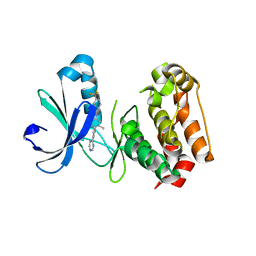 | | Crystal structure of benzamide 9 bound to AuroraA | | Descriptor: | N-{3-methyl-4-[(3-pyrimidin-4-ylpyridin-2-yl)oxy]phenyl}-3-(trifluoromethyl)benzamide, cDNA FLJ58295, highly similar to Serine/threonine-protein kinase 6 | | Authors: | Huang, X. | | Deposit date: | 2010-07-27 | | Release date: | 2010-08-18 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Discovery of a potent, selective, and orally bioavailable pyridinyl-pyrimidine phthalazine aurora kinase inhibitor.
J.Med.Chem., 53, 2010
|
|
3O51
 
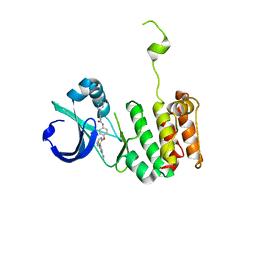 | | Crystal structure of anthranilamide 10 bound to AuroraA | | Descriptor: | N-[4-({3-[5-fluoro-2-(methylideneamino)pyrimidin-4-yl]pyridin-2-yl}oxy)phenyl]-2-(phenylamino)benzamide, cDNA FLJ58295, highly similar to Serine/threonine-protein kinase 6 | | Authors: | Huang, X. | | Deposit date: | 2010-07-27 | | Release date: | 2010-08-18 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Discovery of a potent, selective, and orally bioavailable pyridinyl-pyrimidine phthalazine aurora kinase inhibitor.
J.Med.Chem., 53, 2010
|
|
4L03
 
 | |
5YIS
 
 | | Crystal Structure of AnkB LIR/LC3B complex | | Descriptor: | Ankyrin-2, GLYCEROL, Microtubule-associated proteins 1A/1B light chain 3B, ... | | Authors: | Li, J, Zhu, R, Chen, K, Zheng, H, Yuan, C, Zhang, H, Wang, C, Zhang, M. | | Deposit date: | 2017-10-06 | | Release date: | 2018-05-23 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.201 Å) | | Cite: | Potent and specific Atg8-targeting autophagy inhibitory peptides from giant ankyrins.
Nat. Chem. Biol., 14, 2018
|
|
5YIQ
 
 | | Crystal structure of AnkG LIR/LC3B complex | | Descriptor: | Ankyrin-3, Microtubule-associated proteins 1A/1B light chain 3B, ZINC ION | | Authors: | Li, J, Zhu, R, Chen, K, Zheng, H, Yuan, C, Zhang, H, Wang, C, Zhang, M. | | Deposit date: | 2017-10-06 | | Release date: | 2018-05-23 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Potent and specific Atg8-targeting autophagy inhibitory peptides from giant ankyrins.
Nat. Chem. Biol., 14, 2018
|
|
5YIR
 
 | | Crystal Structure of AnkB LIR/GABARAP complex | | Descriptor: | Ankyrin-2, Gamma-aminobutyric acid receptor-associated protein, NICKEL (II) ION | | Authors: | Li, J, Zhu, R, Chen, K, Zheng, H, Yuan, C, Zhang, H, Wang, C, Zhang, M. | | Deposit date: | 2017-10-06 | | Release date: | 2018-05-23 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.75 Å) | | Cite: | Potent and specific Atg8-targeting autophagy inhibitory peptides from giant ankyrins.
Nat. Chem. Biol., 14, 2018
|
|
5YIP
 
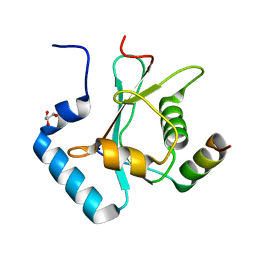 | | Crystal Structure of AnkG LIR/GABARAPL1 complex | | Descriptor: | Ankyrin-3, GLYCEROL, Gamma-aminobutyric acid receptor-associated protein-like 1 | | Authors: | Li, J, Zhu, R, Chen, K, Zheng, H, Yuan, C, Zhang, H, Wang, C, Zhang, M. | | Deposit date: | 2017-10-06 | | Release date: | 2018-05-23 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Potent and specific Atg8-targeting autophagy inhibitory peptides from giant ankyrins.
Nat. Chem. Biol., 14, 2018
|
|
3AME
 
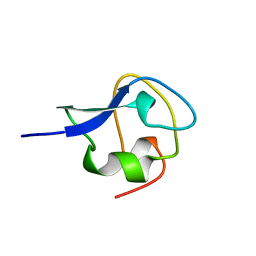 | | TYPE III ANTIFREEZE PROTEIN ISOFORM HPLC 12 Q9TQ44T | | Descriptor: | PROTEIN (ANTIFREEZE PROTEIN TYPE III) | | Authors: | Graether, S.P, Deluca, C.I, Baardsnes, J, Hill, G.A, Davies, P.L, Jia, Z. | | Deposit date: | 1999-01-24 | | Release date: | 1999-04-29 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Quantitative and qualitative analysis of type III antifreeze protein structure and function.
J.Biol.Chem., 274, 1999
|
|
3CHH
 
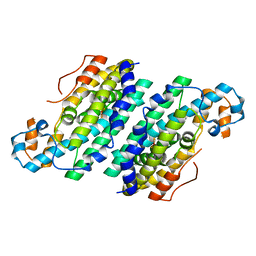 | | Crystal Structure of Di-iron AurF | | Descriptor: | MU-OXO-DIIRON, p-Aminobenzoate N-Oxygenase | | Authors: | Zhang, H, Brunzelle, J.S, Nair, S.K. | | Deposit date: | 2008-03-09 | | Release date: | 2008-05-27 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | In vitro reconstitution and crystal structure of p-aminobenzoate N-oxygenase (AurF) involved in aureothin biosynthesis.
Proc.Natl.Acad.Sci.Usa, 105, 2008
|
|
3CHI
 
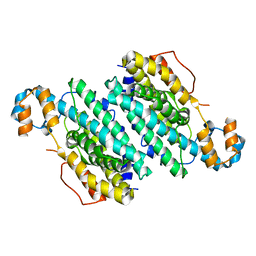 | |
3CHU
 
 | | Crystal Structure of Di-iron Aurf | | Descriptor: | MU-OXO-DIIRON, p-Aminobenzoate N-Oxygenase | | Authors: | Zhang, H, Brunzelle, J.S, Nair, S.K. | | Deposit date: | 2008-03-10 | | Release date: | 2008-05-27 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | In vitro reconstitution and crystal structure of p-aminobenzoate N-oxygenase (AurF) involved in aureothin biosynthesis.
Proc.Natl.Acad.Sci.Usa, 105, 2008
|
|
4HGS
 
 | | Crystal structure of ck1gs with compound 13 | | Descriptor: | 2-{2-[(3,4-difluorophenoxy)methyl]-5-methoxypyridin-4-yl}-1,5,6,7-tetrahydro-4H-pyrrolo[3,2-c]pyridin-4-one, Casein kinase I isoform gamma-3 | | Authors: | Huang, X. | | Deposit date: | 2012-10-08 | | Release date: | 2012-11-21 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Crystal structure of ck1gs with compound 13
Acs Med.Chem.Lett
|
|
4G59
 
 | |
3RDN
 
 | | NMR STRUCTURE OF THE N-TERMINAL DOMAIN WITH A LINKER PORTION OF ANTARCTIC EEL POUT ANTIFREEZE PROTEIN RD3, MINIMIZED AVERAGE STRUCTURE | | Descriptor: | ANTIFREEZE PROTEIN RD3 TYPE III | | Authors: | Miura, K, Ohgiya, S, Hoshino, T, Nemoto, N, Hikichi, K, Tsuda, S. | | Deposit date: | 1998-02-24 | | Release date: | 1999-02-23 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Structural basis for the binding of a globular antifreeze protein to ice.
Nature, 384, 1996
|
|
7AME
 
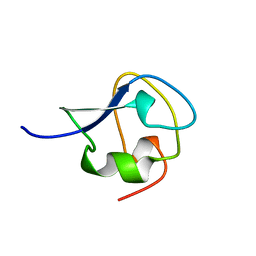 | | TYPE III ANTIFREEZE PROTEIN ISOFORM HPLC 12 T15A | | Descriptor: | PROTEIN (ANTIFREEZE PROTEIN TYPE III) | | Authors: | Graether, S.P, Deluca, C.I, Baardsnes, J, Hill, G.A, Davies, P.L, Jia, Z. | | Deposit date: | 1999-01-24 | | Release date: | 1999-04-29 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Quantitative and qualitative analysis of type III antifreeze protein structure and function.
J.Biol.Chem., 274, 1999
|
|
7SWW
 
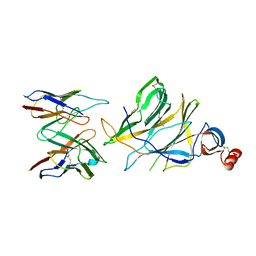 | |
7SWX
 
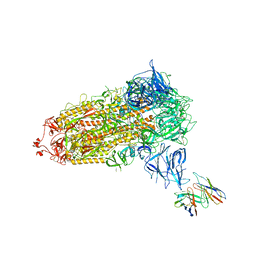 | |
