5JI2
 
 | | HslU L199Q in HslUV complex | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, ATP-dependent protease ATPase subunit HslU, ATP-dependent protease subunit HslV, ... | | Authors: | Grant, R.A, Sauer, R.T, Schmitz, K.R, Baytshtok, V. | | Deposit date: | 2016-04-21 | | Release date: | 2016-11-30 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (3.307 Å) | | Cite: | A Structurally Dynamic Region of the HslU Intermediate Domain Controls Protein Degradation and ATP Hydrolysis.
Structure, 24, 2016
|
|
2PCB
 
 | |
4U01
 
 | | HCV NS3/4A serine protease in complex with 6570 | | Descriptor: | (2S,3aS,10Z,11aS,12aR)-2-({8-fluoro-7-methoxy-2-[4-(propan-2-yl)-1,3-thiazol-2-yl]quinolin-4-yl}oxy)-5-methyl-N-[(1-methylcyclopropyl)sulfonyl]-4,14-dioxo-1,2,3,3a,4,5,6,7,8,9,11a,12,13,14-tetradecahydro-12aH-cyclopropa[m]pyrrolo[1,2-c][1,3,6]triazacyclotetradecine-12a-carboxamide, CHLORIDE ION, NS4A protein, ... | | Authors: | Parsy, C.C, Alexandre, F.-R, Brandt, G, Caillet, C, Chaves, D, Derock, M, Gloux, D, Griffon, Y, Lallos, L.B, Leroy, F, Liuzzi, M, Loi, A.-G, Mayes, B, Moulat, L, Moussa, A, Chiara, M, Roques, V, Rosinovsky, E, Seifer, M, Stewart, A, Wang, J, Standring, D, Surleraux, D. | | Deposit date: | 2014-07-11 | | Release date: | 2015-07-29 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Discovery and structural diversity of the hepatitis C virus NS3/4A serine protease inhibitor series leading to clinical candidate IDX320.
Bioorg.Med.Chem.Lett., 25, 2015
|
|
2PCC
 
 | | CRYSTAL STRUCTURE OF A COMPLEX BETWEEN ELECTRON TRANSFER PARTNERS, CYTOCHROME C PEROXIDASE AND CYTOCHROME C | | Descriptor: | CYTOCHROME C PEROXIDASE, ISO-1-CYTOCHROME C, PROTOPORPHYRIN IX CONTAINING FE, ... | | Authors: | Pelletier, H, Kraut, J. | | Deposit date: | 1993-04-14 | | Release date: | 1993-07-15 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystal structure of a complex between electron transfer partners, cytochrome c peroxidase and cytochrome c.
Science, 258, 1992
|
|
7FEC
 
 | |
7FED
 
 | |
7FEB
 
 | |
1NYJ
 
 | |
7D7L
 
 | | The crystal structure of SARS-CoV-2 papain-like protease in complex with YM155 | | Descriptor: | 1-(2-methoxyethyl)-2-methyl-3-(pyrazin-2-ylmethyl)benzo[f]benzimidazol-3-ium-4,9-dione, CAFFEINE, GLYCEROL, ... | | Authors: | Zhao, Y, Sun, L, Yang, H.T, Rao, Z.H. | | Deposit date: | 2020-10-04 | | Release date: | 2021-04-21 | | Last modified: | 2021-11-17 | | Method: | X-RAY DIFFRACTION (2.11 Å) | | Cite: | High-throughput screening identifies established drugs as SARS-CoV-2 PLpro inhibitors.
Protein Cell, 12, 2021
|
|
7D7K
 
 | | The crystal structure of SARS-CoV-2 papain-like protease in apo form | | Descriptor: | 1,2-ETHANEDIOL, CAFFEINE, Non-structural protein 3, ... | | Authors: | Zhao, Y, Sun, L, Yang, H.T, Rao, Z.H. | | Deposit date: | 2020-10-04 | | Release date: | 2021-04-21 | | Last modified: | 2021-11-17 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | High-throughput screening identifies established drugs as SARS-CoV-2 PLpro inhibitors.
Protein Cell, 12, 2021
|
|
7DFE
 
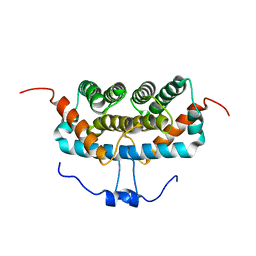 | | NMR structure of TuSp2-RP | | Descriptor: | B6 protein | | Authors: | Lin, Z, Fan, T, Fan, J. | | Deposit date: | 2020-11-07 | | Release date: | 2021-11-17 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | 1H, 15N and 13C resonance assignments of a repetitive domain of tubuliform spidroin 2
Biomol.Nmr Assign., 15, 2021
|
|
7DFB
 
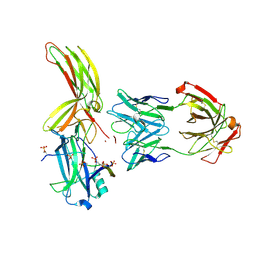 | | Crystal of Arrestin2-V2Rpp-6-7-Fab30 complex | | Descriptor: | Beta-arrestin-1, FAB30 HEAVY CHAIN, FAB30 LIGHT CHAIN, ... | | Authors: | Sun, J.P, Yu, X, Xiao, P, He, Q.T, Lin, J.Y, Zhu, Z.L. | | Deposit date: | 2020-11-06 | | Release date: | 2021-07-28 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (3.28 Å) | | Cite: | Structural studies of phosphorylation-dependent interactions between the V2R receptor and arrestin-2.
Nat Commun, 12, 2021
|
|
7DFA
 
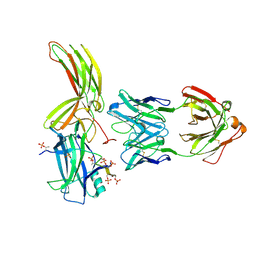 | | Crystal of Arrestin2-V2Rpp-4-Fab30 complex | | Descriptor: | Beta-arrestin-1, FAB30 HEAVY CHAIN, FAB30 LIGHT CHAIN, ... | | Authors: | Sun, J.P, Yu, X, Xiao, P, He, Q.T, Lin, J.Y, Zhu, Z.L. | | Deposit date: | 2020-11-06 | | Release date: | 2021-07-28 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.54 Å) | | Cite: | Structural studies of phosphorylation-dependent interactions between the V2R receptor and arrestin-2.
Nat Commun, 12, 2021
|
|
7DF9
 
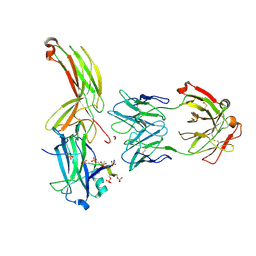 | | Crystal of Arrestin2-V2Rpp-1-Fab30 complex | | Descriptor: | Beta-arrestin-1, FAB30 HEAVY CHAIN, FAB30 LIGHT CHAIN, ... | | Authors: | Sun, J.P, Yu, X, Xiao, P, He, Q.T, Lin, J.Y, Zhu, Z.L. | | Deposit date: | 2020-11-06 | | Release date: | 2021-07-28 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (3.17 Å) | | Cite: | Structural studies of phosphorylation-dependent interactions between the V2R receptor and arrestin-2.
Nat Commun, 12, 2021
|
|
7DFC
 
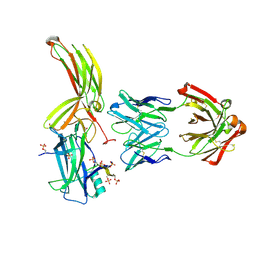 | | Crystal of Arrestin2-V2Rpp-3-Fab30 complex | | Descriptor: | Beta-arrestin-1, FAB30 HEAVY CHAIN, FAB30 LIGHT CHAIN, ... | | Authors: | Sun, J.P, Yu, X, Xiao, P, He, Q.T, Lin, J.Y, Zhu, Z.L. | | Deposit date: | 2020-11-06 | | Release date: | 2021-07-28 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.49 Å) | | Cite: | Structural studies of phosphorylation-dependent interactions between the V2R receptor and arrestin-2.
Nat Commun, 12, 2021
|
|
4J2B
 
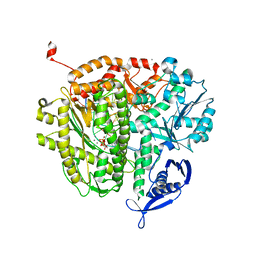 | | RB69 DNA Polymerase L415G Ternary Complex | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, CALCIUM ION, DNA (5'-D(*GP*CP*GP*GP*AP*CP*TP*GP*CP*TP*TP*AP*T)-3'), ... | | Authors: | Xia, S, Wang, J, Konigsberg, W.H. | | Deposit date: | 2013-02-04 | | Release date: | 2014-02-19 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.04 Å) | | Cite: | Alteration in the cavity size adjacent to the active site of RB69 DNA polymerase changes its conformational dynamics.
Nucleic Acids Res., 41, 2013
|
|
7EAG
 
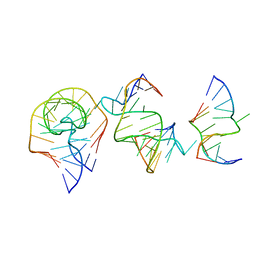 | | Crystal structure of the RAGATH-18 k-turn | | Descriptor: | RNA (5'-R(*GP*UP*CP*UP*AP*UP*GP*AP*AP*GP*GP*CP*UP*GP*GP*AP*GP*AP*C)-3') | | Authors: | Huang, L, Lilley, D.M.J. | | Deposit date: | 2021-03-07 | | Release date: | 2021-06-02 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structure and folding of four putative kink turns identified in structured RNA species in a test of structural prediction rules.
Nucleic Acids Res., 49, 2021
|
|
7EAF
 
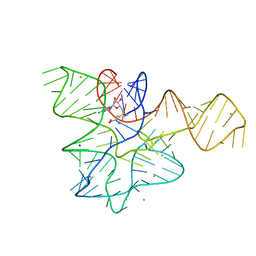 | |
7EK6
 
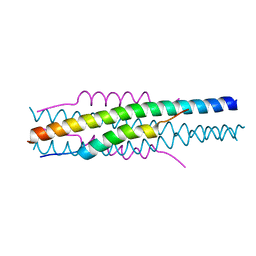 | | Structure of viral peptides IPB19/N52 | | Descriptor: | Spike protein S2 | | Authors: | Yu, D, Qin, B, Cui, S, He, Y. | | Deposit date: | 2021-04-04 | | Release date: | 2021-06-09 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.243 Å) | | Cite: | Structure-based design and characterization of novel fusion-inhibitory lipopeptides against SARS-CoV-2 and emerging variants.
Emerg Microbes Infect, 10, 2021
|
|
7CFG
 
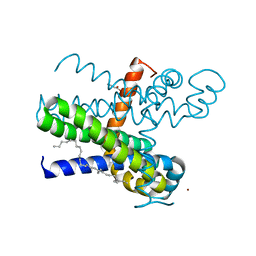 | | Structure of the transmembrane domain of the bacterial CNNM/CorC family Mg2+ transporter in complex with Mg2+ | | Descriptor: | (2R)-2,3-dihydroxypropyl (9Z)-octadec-9-enoate, Hemolysin, MAGNESIUM ION, ... | | Authors: | Huang, Y, Jin, F, Hattori, M. | | Deposit date: | 2020-06-25 | | Release date: | 2021-02-24 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Structural basis for the Mg 2+ recognition and regulation of the CorC Mg 2+ transporter.
Sci Adv, 7, 2021
|
|
7CFF
 
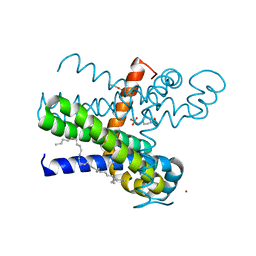 | | Structure of the thermostabilized transmembrane domain of the bacterial CNNM/CorC family Mg2+ transporter in complex with Mg2+ | | Descriptor: | (2R)-2,3-dihydroxypropyl (9Z)-octadec-9-enoate, 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, Hemolysin, ... | | Authors: | Huang, Y, Jin, F, Hattori, M. | | Deposit date: | 2020-06-25 | | Release date: | 2021-02-24 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural basis for the Mg 2+ recognition and regulation of the CorC Mg 2+ transporter.
Sci Adv, 7, 2021
|
|
7CFH
 
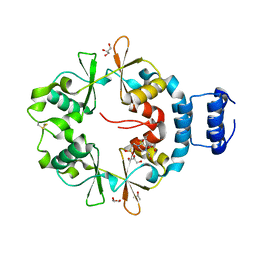 | |
7CFI
 
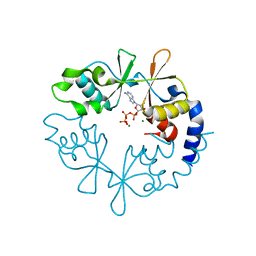 | |
7CHU
 
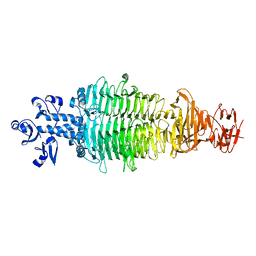 | | Geobacillus virus E2 - ORF18 | | Descriptor: | Putative pectin lyase | | Authors: | Gong, Y. | | Deposit date: | 2020-07-06 | | Release date: | 2021-04-14 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (2.008 Å) | | Cite: | Structural and functional characterization of the deep-sea thermophilic bacteriophage GVE2 tailspike protein.
Int.J.Biol.Macromol., 164, 2020
|
|
7FEW
 
 | |
