8OWW
 
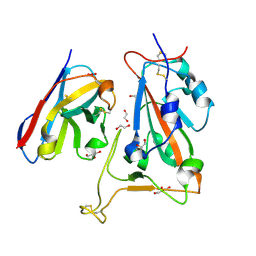 | | B5-5 nanobody bound to SARS-CoV-2 spike RBD (Wuhan) | | Descriptor: | 1,2-ETHANEDIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose, B5-5 nanobody, ... | | Authors: | Cornish, K.A.S, Naismith, J.H, Owens, R.J. | | Deposit date: | 2023-04-28 | | Release date: | 2024-05-08 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.969 Å) | | Cite: | Structural and functional characterization of nanobodies that neutralize Omicron variants of SARS-CoV-2.
Open Biology, 14, 2024
|
|
8OWV
 
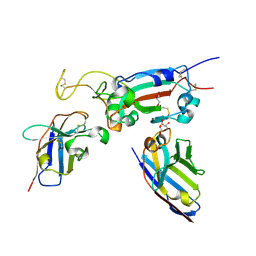 | | H6 and F2 nanobodies bound to SARS-CoV-2 spike RBD | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, F2, GLYCEROL, ... | | Authors: | Mikolajek, H, Naismith, J.H, Owens, R.J. | | Deposit date: | 2023-04-28 | | Release date: | 2024-05-08 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (1.73 Å) | | Cite: | Structural and functional characterization of nanobodies that neutralize Omicron variants of SARS-CoV-2.
Open Biology, 14, 2024
|
|
8OYU
 
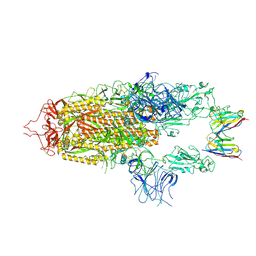 | | Stabilised BA.1 SARS-CoV-2 spike with H6 nanobodies in '2 up 1 down' RBD conformation | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, H6 nanobody, ... | | Authors: | Weckener, M, Naismith, J.H, Owens, R.J. | | Deposit date: | 2023-05-05 | | Release date: | 2024-05-15 | | Last modified: | 2024-10-16 | | Method: | ELECTRON MICROSCOPY (4 Å) | | Cite: | Structural and functional characterization of nanobodies that neutralize Omicron variants of SARS-CoV-2.
Open Biology, 14, 2024
|
|
3DKS
 
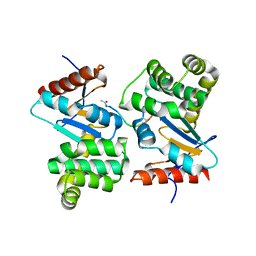 | | DsbA substrate complex | | Descriptor: | Thiol:disulfide interchange protein dsbA, siga peptide | | Authors: | Paxman, J.J, Borg, N.A, Horne, J, Rossjohn, J, Thompson, P.E, Piek, S, Kahler, C.M, Sakellaris, H, Scanlon, M.J. | | Deposit date: | 2008-06-25 | | Release date: | 2009-05-12 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | The structure of the bacterial oxidoreductase enzyme DsbA in complex with a peptide reveals a basis for substrate specificity in the catalytic cycle of DsbA enzymes
J.Biol.Chem., 284, 2009
|
|
8I7J
 
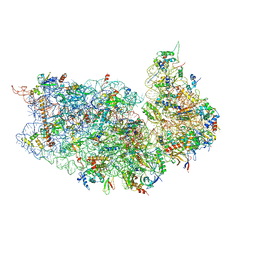 | |
8T8P
 
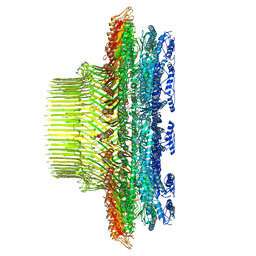 | | 33-mer FliF MS-ring from Salmonella | | Descriptor: | Flagellar M-ring protein | | Authors: | Singh, P.K, Iverson, T.M. | | Deposit date: | 2023-06-23 | | Release date: | 2024-02-28 | | Last modified: | 2024-08-21 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | CryoEM structures reveal how the bacterial flagellum rotates and switches direction.
Nat Microbiol, 9, 2024
|
|
8SWH
 
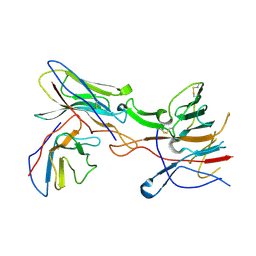 | |
8VKR
 
 | | CW Flagellar Switch Complex with extra density - FliF, FliG, FliM, and FliN forming the C-ring from Salmonella | | Descriptor: | Flagellar M-ring protein, Flagellar motor switch protein FliG, Flagellar motor switch protein FliM, ... | | Authors: | Singh, P.K, Iverson, T.M. | | Deposit date: | 2024-01-09 | | Release date: | 2024-02-28 | | Last modified: | 2024-08-21 | | Method: | ELECTRON MICROSCOPY (5.9 Å) | | Cite: | CryoEM structures reveal how the bacterial flagellum rotates and switches direction.
Nat Microbiol, 9, 2024
|
|
8VID
 
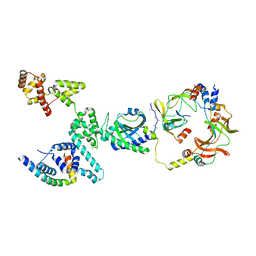 | | CW Flagellar Switch Complex with extra density - FliF, FliG, FliM, and FliN forming single subunit of the C-ring from Salmonella | | Descriptor: | Flagellar M-ring protein, Flagellar motor switch protein FliG, Flagellar motor switch protein FliM, ... | | Authors: | Singh, P.K, Iverson, T.M. | | Deposit date: | 2024-01-03 | | Release date: | 2024-02-28 | | Last modified: | 2024-08-21 | | Method: | ELECTRON MICROSCOPY (5.9 Å) | | Cite: | CryoEM structures reveal how the bacterial flagellum rotates and switches direction.
Nat Microbiol, 9, 2024
|
|
8VIB
 
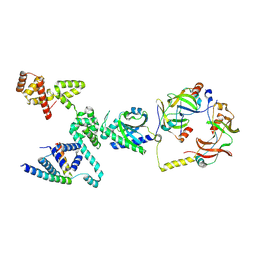 | | CW Flagellar Switch Complex - FliF, FliG, FliM, and FliN forming single subunit of the C-ring from Salmonella | | Descriptor: | Flagellar M-ring protein, Flagellar motor switch protein FliG, Flagellar motor switch protein FliM, ... | | Authors: | Singh, P.K, Iverson, T.M. | | Deposit date: | 2024-01-03 | | Release date: | 2024-02-28 | | Last modified: | 2024-08-21 | | Method: | ELECTRON MICROSCOPY (4.6 Å) | | Cite: | CryoEM structures reveal how the bacterial flagellum rotates and switches direction.
Nat Microbiol, 9, 2024
|
|
8VKQ
 
 | | CW Flagellar Switch Complex - FliF, FliG, FliM, and FliN forming the C-ring from Salmonella | | Descriptor: | Flagellar M-ring protein, Flagellar motor switch protein FliG, Flagellar motor switch protein FliM, ... | | Authors: | Singh, P.K, Iverson, T.M. | | Deposit date: | 2024-01-09 | | Release date: | 2024-02-28 | | Last modified: | 2024-08-21 | | Method: | ELECTRON MICROSCOPY (4.6 Å) | | Cite: | CryoEM structures reveal how the bacterial flagellum rotates and switches direction.
Nat Microbiol, 9, 2024
|
|
4DVC
 
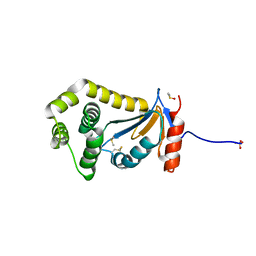 | | Structural and functional studies of TcpG, the Vibrio cholerae DsbA disulfide-forming protein required for pilus and cholera toxin production | | Descriptor: | DIMETHYL SULFOXIDE, SULFATE ION, Thiol:disulfide interchange protein DsbA | | Authors: | Walden, P.M, Martin, J.L. | | Deposit date: | 2012-02-23 | | Release date: | 2012-10-31 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (1.2 Å) | | Cite: | The 1.2 A resolution crystal structure of TcpG, the Vibrio cholerae DsbA disulfide-forming protein required for pilus and cholera-toxin production
Acta Crystallogr.,Sect.D, 68, 2012
|
|
6SRV
 
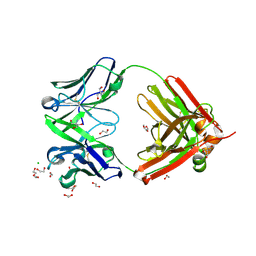 | | Structure of the arginase-2-inhibitory human antigen-binding fragment Fab C0021144 | | Descriptor: | 1,2-ETHANEDIOL, 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, CHLORIDE ION, ... | | Authors: | Burschowsky, D, Addyman, A, Fiedler, S, Groves, M, Haynes, S, Seewooruthun, C, Carr, M. | | Deposit date: | 2019-09-06 | | Release date: | 2020-06-10 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structural and functional characterization of C0021158, a high-affinity monoclonal antibody that inhibits Arginase 2 function via a novel non-competitive mechanism of action.
Mabs, 12
|
|
6TUL
 
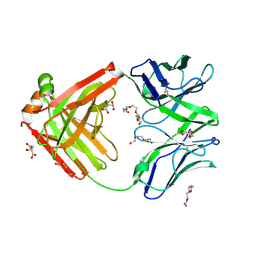 | | Structure of the arginase-2-inhibitory human antigen-binding fragment Fab C0021177 | | Descriptor: | D-MALATE, Fab C0021177 heavy chain (IgG1), Fab C0021177 light chain (IgG1), ... | | Authors: | Burschowsky, D, Addyman, A, Fiedler, S, Groves, M, Haynes, S, Seewooruthun, C, Carr, M. | | Deposit date: | 2020-01-07 | | Release date: | 2020-06-10 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Structural and functional characterization of C0021158, a high-affinity monoclonal antibody that inhibits Arginase 2 function via a novel non-competitive mechanism of action.
Mabs, 12
|
|
6SRX
 
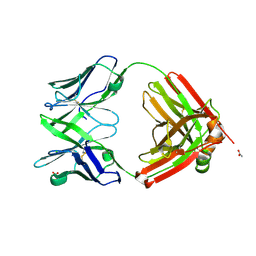 | | Structure of the arginase-2-inhibitory human antigen-binding fragment Fab C0021158 | | Descriptor: | ACETATE ION, CHLORIDE ION, Fab C0021158 heavy chain (IgG1), ... | | Authors: | Burschowsky, D, Addyman, A, Fiedler, S, Groves, M, Haynes, S, Seewooruthun, C, Carr, M. | | Deposit date: | 2019-09-06 | | Release date: | 2020-06-10 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structural and functional characterization of C0021158, a high-affinity monoclonal antibody that inhibits Arginase 2 function via a novel non-competitive mechanism of action.
Mabs, 12
|
|
6SS0
 
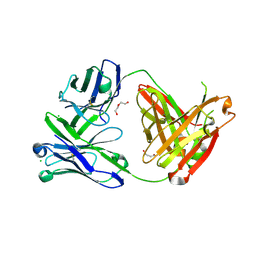 | | Structure of the arginase-2-inhibitory human antigen-binding fragment Fab C0021181 | | Descriptor: | CHLORIDE ION, DI(HYDROXYETHYL)ETHER, Fab C0021181 heavy chain (IgG1), ... | | Authors: | Burschowsky, D, Addyman, A, Fiedler, S, Groves, M, Haynes, S, Seewooruthun, C, Carr, M. | | Deposit date: | 2019-09-06 | | Release date: | 2020-06-10 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structural and functional characterization of C0021158, a high-affinity monoclonal antibody that inhibits Arginase 2 function via a novel non-competitive mechanism of action.
Mabs, 12
|
|
7S1C
 
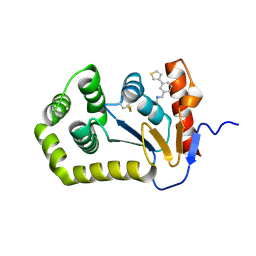 | | Crystal structure of E.coli DsbA in complex with compound MIPS-0001897 (compound 1) | | Descriptor: | COPPER (II) ION, Thiol:disulfide interchange protein DsbA, ~{N}-methyl-1-(3-thiophen-3-ylphenyl)methanamine | | Authors: | Heras, B, Scanlon, M.J, Martin, J.L, Sharma, P. | | Deposit date: | 2021-09-02 | | Release date: | 2023-02-08 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.949 Å) | | Cite: | Fluoromethylketone-fragment conjugates designed as covalent modifiers of EcDsbA are atypical substrates
Chemrxiv, 2022
|
|
4MWI
 
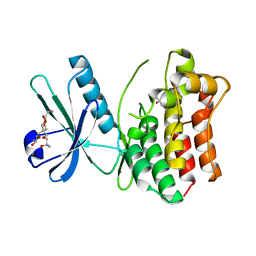 | | Crystal structure of the human MLKL pseudokinase domain | | Descriptor: | (2S)-1-[3-{[(2R)-2-hydroxypropyl]oxy}-2,2-bis({[(2R)-2-hydroxypropyl]oxy}methyl)propoxy]propan-2-ol, GLYCEROL, Mixed lineage kinase domain-like protein | | Authors: | Czabotar, P.E, Murphy, J.M. | | Deposit date: | 2013-09-25 | | Release date: | 2013-12-04 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Insights into the evolution of divergent nucleotide-binding mechanisms among pseudokinases revealed by crystal structures of human and mouse MLKL.
Biochem.J., 457, 2014
|
|
5OBR
 
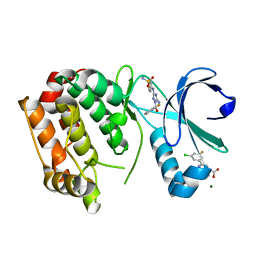 | | Aurora A kinase in complex with 2-(3-chloro-5-fluorophenyl)quinoline-4-carboxylic acid and JNJ-7706621 | | Descriptor: | 2-(3-chloranyl-5-fluoranyl-phenyl)quinoline-4-carboxylic acid, 4-({5-amino-1-[(2,6-difluorophenyl)carbonyl]-1H-1,2,4-triazol-3-yl}amino)benzenesulfonamide, Aurora kinase A, ... | | Authors: | Rossmann, M, Janecek, M, Hyvonen, M. | | Deposit date: | 2017-06-29 | | Release date: | 2017-08-09 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.62 Å) | | Cite: | Computationally-guided optimization of small-molecule inhibitors of the Aurora A kinase-TPX2 protein-protein interaction.
Chem. Commun. (Camb.), 53, 2017
|
|
5OBJ
 
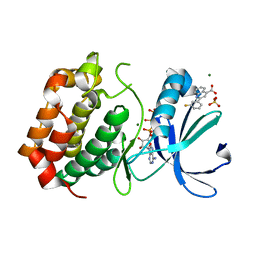 | | Aurora A kinase in complex with 2-(3-fluorophenyl)quinoline-4-carboxylic acid and ATP | | Descriptor: | 2-(3-fluorophenyl)quinoline-4-carboxylic acid, ADENOSINE-5'-TRIPHOSPHATE, Aurora kinase A, ... | | Authors: | Rossmann, M, Janecek, M, Hyvonen, M. | | Deposit date: | 2017-06-28 | | Release date: | 2017-08-09 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.901 Å) | | Cite: | Computationally-guided optimization of small-molecule inhibitors of the Aurora A kinase-TPX2 protein-protein interaction.
Chem. Commun. (Camb.), 53, 2017
|
|
