8K14
 
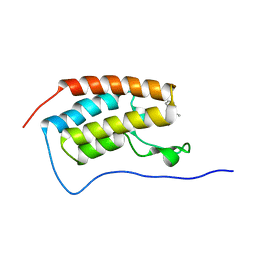 | | X-ray crystal structure of 18a in BRD4(1) | | Descriptor: | 4-[8-methoxy-2-methyl-1-(1-phenylethyl)imidazo[4,5-c]quinolin-7-yl]-3,5-dimethyl-1,2-oxazole, Bromodomain-containing protein 4 | | Authors: | Xu, H, Shen, H, Zhang, Y, Xu, Y, Li, R. | | Deposit date: | 2023-07-10 | | Release date: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (1.28 Å) | | Cite: | Discovery of (R)-4-(8-methoxy-2-methyl-1-(1-phenylethy)-1H-imidazo[4,5-c]quinnolin-7-yl)-3,5-dimethylisoxazole as a potent and selective BET inhibitor for treatment of acute myeloid leukemia (AML) guided by FEP calculation.
Eur.J.Med.Chem., 263, 2024
|
|
8E0Q
 
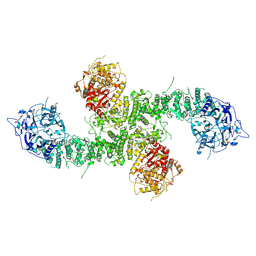 | | Structure of the human UBR5 HECT-type E3 ubiquitin ligase in a C2 symmetric dimeric form | | Descriptor: | E3 ubiquitin-protein ligase UBR5, ZINC ION | | Authors: | Wang, F, He, Q, Lin, G, Li, H. | | Deposit date: | 2022-08-09 | | Release date: | 2023-04-19 | | Last modified: | 2024-06-12 | | Method: | ELECTRON MICROSCOPY (2.66 Å) | | Cite: | Structure of the human UBR5 E3 ubiquitin ligase.
Structure, 31, 2023
|
|
7DW9
 
 | |
7EO0
 
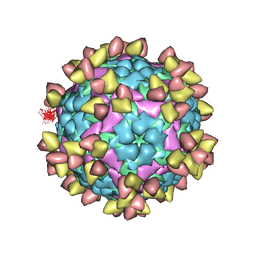 | | FOOT AND MOUTH DISEASE VIRUS O/TIBET/99-BOUND THE SINGLE CHAIN FRAGMEN ANTIBODY C4 | | Descriptor: | Ig heavy chain variable region, Ig lamda chain variable region, O/TIBET/99 VP1, ... | | Authors: | He, Y, Li, K. | | Deposit date: | 2021-04-21 | | Release date: | 2021-08-18 | | Last modified: | 2024-10-09 | | Method: | ELECTRON MICROSCOPY (3.75 Å) | | Cite: | Two Cross-Protective Antigen Sites on Foot-and-Mouth Disease Virus Serotype O Structurally Revealed by Broadly Neutralizing Antibodies from Cattle.
J.Virol., 95, 2021
|
|
7E4M
 
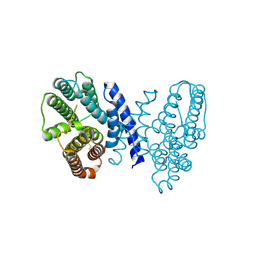 | |
7E4O
 
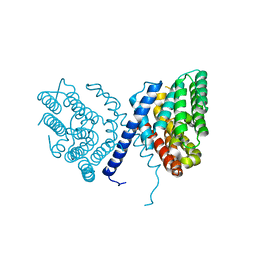 | |
7E4N
 
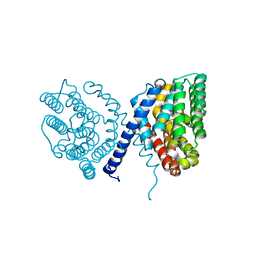 | | Crystal structure of Sat1646 | | Descriptor: | MAGNESIUM ION, Sat1646 | | Authors: | Yu, J.H, Xing, B.Y, Ma, M. | | Deposit date: | 2021-02-14 | | Release date: | 2021-10-06 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (1.94 Å) | | Cite: | Functional characterization and structural bases of two class I diterpene synthases in pimarane-type diterpene biosynthesis
Commun Chem, 4, 2021
|
|
8EWI
 
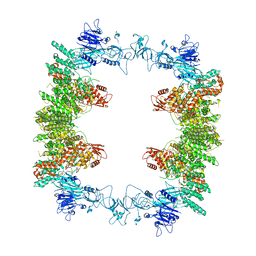 | | Structure of the human UBR5 HECT-type E3 ubiquitin ligase in a tetrameric form | | Descriptor: | E3 ubiquitin-protein ligase UBR5, ZINC ION | | Authors: | Wang, F, He, Q, Lin, G, Li, H. | | Deposit date: | 2022-10-23 | | Release date: | 2023-04-19 | | Last modified: | 2024-06-19 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | Structure of the human UBR5 E3 ubiquitin ligase.
Structure, 31, 2023
|
|
5UTT
 
 | | SrtA sortase from Actinomyces oris | | Descriptor: | CHLORIDE ION, Sortase | | Authors: | Osipiuk, J, Ma, X, Ton-That, H, Anderson, W.F, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2017-02-15 | | Release date: | 2017-03-01 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Cell-to-cell interaction requires optimal positioning of a pilus tip adhesin modulated by gram-positive transpeptidase enzymes.
Proc.Natl.Acad.Sci.USA, 116, 2019
|
|
5I8E
 
 | | Crystal Structure of Broadly Neutralizing HIV-1 Fusion Peptide-Targeting Antibody VRC34.01 Fab | | Descriptor: | VRC34.01 Fab heavy chain, VRC34.01 Fab light chain, ZINC ION | | Authors: | Xu, K, Zhou, T, Liu, K, Kwong, P.D. | | Deposit date: | 2016-02-18 | | Release date: | 2016-05-25 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.655 Å) | | Cite: | Fusion peptide of HIV-1 as a site of vulnerability to neutralizing antibody.
Science, 352, 2016
|
|
5I8C
 
 | | Crystal Structure of HIV-1 Clade A BG505 Fusion Peptide (residue 512-520) in Complex with Broadly Neutralizing Antibody VRC34.01 Fab | | Descriptor: | HIV-1 Clade A BG505 Fusion Peptide (residue 512-520), VRC34.01 Fab heavy chain, VRC34.01 Fab light chain | | Authors: | Xu, K, Zhou, T, Liu, K, Kwong, P.D. | | Deposit date: | 2016-02-18 | | Release date: | 2016-05-25 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.54 Å) | | Cite: | Fusion peptide of HIV-1 as a site of vulnerability to neutralizing antibody.
Science, 352, 2016
|
|
8X5D
 
 | |
7T79
 
 | |
7T78
 
 | |
8I3E
 
 | |
8HJF
 
 | | Crystal structure of glycosyltransferase SgUGT94-289-3 in complex with M5, state 2 | | Descriptor: | (20S)-2,5,8,11,14,17-HEXAMETHYL-3,6,9,12,15,18-HEXAOXAHENICOSANE-1,20-DIOL, (2S,3S,4S,5R,6R)-2-(hydroxymethyl)-6-[[(2R,3S,4S,5R,6R)-6-[[(3S,8S,9R,10R,11R,13R,14S,17R)-17-[(2S,5R)-5-[(2S,3R,4S,5S,6R)-3-[(2R,3R,4S,5S,6S)-6-(hydroxymethyl)-3,4,5-tris(oxidanyl)oxan-2-yl]oxy-6-[[(2R,3R,4S,5S,6S)-6-(hydroxymethyl)-3,4,5-tris(oxidanyl)oxan-2-yl]oxymethyl]-4,5-bis(oxidanyl)oxan-2-yl]oxy-6-methyl-6-oxidanyl-heptan-2-yl]-4,4,9,13,14-pentamethyl-11-oxidanyl-2,3,7,8,10,11,12,15,16,17-decahydro-1H-cyclopenta[a]phenanthren-3-yl]oxy]-3,4,5-tris(oxidanyl)oxan-2-yl]methoxy]oxane-3,4,5-triol, 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, ... | | Authors: | Li, M, Zhang, S, Cui, S. | | Deposit date: | 2022-11-23 | | Release date: | 2024-05-29 | | Last modified: | 2024-08-14 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural insights into the catalytic selectivity of glycosyltransferase SgUGT94-289-3 towards mogrosides.
Nat Commun, 15, 2024
|
|
8HJP
 
 | | Crystal structure of glycosyltransferase SgUGT94-289-3 in complex with UDP state 1 | | Descriptor: | (20S)-2,5,8,11,14,17-HEXAMETHYL-3,6,9,12,15,18-HEXAOXAHENICOSANE-1,20-DIOL, URIDINE-5'-DIPHOSPHATE, glycosyltranseferease | | Authors: | Li, M, Zhang, S, Cui, S. | | Deposit date: | 2022-11-23 | | Release date: | 2024-05-29 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structural insights into the catalytic selectivity of glycosyltransferase SgUGT94-289-3 towards mogrosides.
Nat Commun, 15, 2024
|
|
8HJN
 
 | |
8HJO
 
 | | Crystal structure of glycosyltransferase SgUGT94-289-3 in complex with UDP state 2 | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, DI(HYDROXYETHYL)ETHER, URIDINE-5'-DIPHOSPHATE, ... | | Authors: | Li, M, Zhang, S, Cui, S. | | Deposit date: | 2022-11-23 | | Release date: | 2024-05-29 | | Last modified: | 2024-08-14 | | Method: | X-RAY DIFFRACTION (1.64 Å) | | Cite: | Structural insights into the catalytic selectivity of glycosyltransferase SgUGT94-289-3 towards mogrosides.
Nat Commun, 15, 2024
|
|
8HJG
 
 | | Crystal structure of glycosyltransferase SgUGT94-289-3 in complex with M5, state 1 | | Descriptor: | (20S)-2,5,8,11,14,17-HEXAMETHYL-3,6,9,12,15,18-HEXAOXAHENICOSANE-1,20-DIOL, (2S,3S,4S,5R,6R)-2-(hydroxymethyl)-6-[[(2R,3S,4S,5R,6R)-6-[[(3S,8S,9R,10R,11R,13R,14S,17R)-17-[(2S,5R)-5-[(2S,3R,4S,5S,6R)-3-[(2R,3R,4S,5S,6S)-6-(hydroxymethyl)-3,4,5-tris(oxidanyl)oxan-2-yl]oxy-6-[[(2R,3R,4S,5S,6S)-6-(hydroxymethyl)-3,4,5-tris(oxidanyl)oxan-2-yl]oxymethyl]-4,5-bis(oxidanyl)oxan-2-yl]oxy-6-methyl-6-oxidanyl-heptan-2-yl]-4,4,9,13,14-pentamethyl-11-oxidanyl-2,3,7,8,10,11,12,15,16,17-decahydro-1H-cyclopenta[a]phenanthren-3-yl]oxy]-3,4,5-tris(oxidanyl)oxan-2-yl]methoxy]oxane-3,4,5-triol, 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, ... | | Authors: | Li, M, Zhang, S, Cui, S. | | Deposit date: | 2022-11-23 | | Release date: | 2024-05-29 | | Last modified: | 2024-08-14 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structural insights into the catalytic selectivity of glycosyltransferase SgUGT94-289-3 towards mogrosides.
Nat Commun, 15, 2024
|
|
8HJQ
 
 | |
8HJL
 
 | | Crystal structure of glycosyltransferase SgUGT94-289-3 in complex with M3E | | Descriptor: | (20S)-2,5,8,11,14,17-HEXAMETHYL-3,6,9,12,15,18-HEXAOXAHENICOSANE-1,20-DIOL, (2R,3S,4S,5R,6R)-2-(hydroxymethyl)-6-[[(3S,8S,9R,10S,11S,13R,14S,17S)-17-[(2S,5R)-5-[(2S,3R,4S,5R,6S)-6-(hydroxymethyl)-3-[(2S,3R,4S,5S,6R)-6-(hydroxymethyl)-3,4,5-tris(oxidanyl)oxan-2-yl]oxy-4,5-bis(oxidanyl)oxan-2-yl]oxy-6-methyl-6-oxidanyl-heptan-2-yl]-4,4,9,13,14-pentamethyl-11-oxidanyl-2,3,7,8,10,11,12,15,16,17-decahydro-1H-cyclopenta[a]phenanthren-3-yl]oxy]oxane-3,4,5-triol, 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, ... | | Authors: | Li, M, Zhang, S, Cui, S. | | Deposit date: | 2022-11-23 | | Release date: | 2024-05-29 | | Last modified: | 2024-08-14 | | Method: | X-RAY DIFFRACTION (1.72 Å) | | Cite: | Structural insights into the catalytic selectivity of glycosyltransferase SgUGT94-289-3 towards mogrosides.
Nat Commun, 15, 2024
|
|
8HJH
 
 | | Crystal structure of glycosyltransferase SgUGT94-289-3 in complex with SIA, state 3 | | Descriptor: | (20S)-2,5,8,11,14,17-HEXAMETHYL-3,6,9,12,15,18-HEXAOXAHENICOSANE-1,20-DIOL, (2S,3S,4S,5R,6R)-2-(hydroxymethyl)-6-[[(2S,3S,4S,5R,6S)-5-[(2S,3R,4S,5S,6S)-6-(hydroxymethyl)-3,4,5-tris(oxidanyl)oxan-2-yl]oxy-6-[(3R,6S)-6-[(3S,8S,9R,10R,11S,13R,14S,17R)-3-[(2S,3R,4S,5S,6S)-6-(hydroxymethyl)-3,4,5-tris(oxidanyl)oxan-2-yl]oxy-4,4,9,13,14-pentamethyl-11-oxidanyl-2,3,7,8,10,11,12,15,16,17-decahydro-1H-cyclopenta[a]phenanthren-17-yl]-2-methyl-2-oxidanyl-heptan-3-yl]oxy-3,4-bis(oxidanyl)oxan-2-yl]methoxy]oxane-3,4,5-triol, 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, ... | | Authors: | Li, M, Zhang, S, Cui, S. | | Deposit date: | 2022-11-23 | | Release date: | 2024-05-29 | | Last modified: | 2024-08-14 | | Method: | X-RAY DIFFRACTION (1.76 Å) | | Cite: | Structural insights into the catalytic selectivity of glycosyltransferase SgUGT94-289-3 towards mogrosides.
Nat Commun, 15, 2024
|
|
8HJK
 
 | | Crystal structure of glycosyltransferase SgUGT94-289-3 in complex with SIA, state 1 | | Descriptor: | (2S,3S,4S,5R,6R)-2-(hydroxymethyl)-6-[[(2S,3S,4S,5R,6S)-5-[(2S,3R,4S,5S,6S)-6-(hydroxymethyl)-3,4,5-tris(oxidanyl)oxan-2-yl]oxy-6-[(3R,6S)-6-[(3S,8S,9R,10R,11S,13R,14S,17R)-3-[(2S,3R,4S,5S,6S)-6-(hydroxymethyl)-3,4,5-tris(oxidanyl)oxan-2-yl]oxy-4,4,9,13,14-pentamethyl-11-oxidanyl-2,3,7,8,10,11,12,15,16,17-decahydro-1H-cyclopenta[a]phenanthren-17-yl]-2-methyl-2-oxidanyl-heptan-3-yl]oxy-3,4-bis(oxidanyl)oxan-2-yl]methoxy]oxane-3,4,5-triol, URIDINE-5'-DIPHOSPHATE, glycosyltranseferease | | Authors: | Li, M, Zhang, S, Cui, S. | | Deposit date: | 2022-11-23 | | Release date: | 2024-05-29 | | Last modified: | 2024-08-14 | | Method: | X-RAY DIFFRACTION (1.833 Å) | | Cite: | Structural insights into the catalytic selectivity of glycosyltransferase SgUGT94-289-3 towards mogrosides.
Nat Commun, 15, 2024
|
|
7WVM
 
 | | The complex structure of PD-1 and cemiplimab | | Descriptor: | Heavy Chain of Cemiplimab, Light Chain of Cemiplimab, Programmed cell death protein 1 | | Authors: | Lu, D, Xu, Z.P, Liu, K.F, Tan, S.G, Gao, G.F, Chai, Y. | | Deposit date: | 2022-02-10 | | Release date: | 2022-04-20 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (3.4 Å) | | Cite: | PD-1 N58-Glycosylation-Dependent Binding of Monoclonal Antibody Cemiplimab for Immune Checkpoint Therapy.
Front Immunol, 13, 2022
|
|
