4ENM
 
 | |
4ENK
 
 | |
4HDV
 
 | |
4ENN
 
 | |
4ENJ
 
 | |
4HDU
 
 | |
3BJE
 
 | |
1GYQ
 
 | |
1JQY
 
 | | HEAT-LABILE ENTEROTOXIN B-PENTAMER WITH LIGAND BMSC-0010 | | Descriptor: | (3-NITRO-5-(3-MORPHOLIN-4-YL-PROPYLAMINOCARBONYL)PHENYL)-GALACTOPYRANOSIDE, HEAT-LABILE ENTEROTOXIN B CHAIN | | Authors: | Merritt, E.A, Hol, W.G.J. | | Deposit date: | 2001-08-09 | | Release date: | 2002-05-08 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.14 Å) | | Cite: | Anchor-based design of improved cholera toxin and E. coli heat-labile enterotoxin receptor binding antagonists that display multiple binding modes.
Chem.Biol., 9, 2002
|
|
2B4G
 
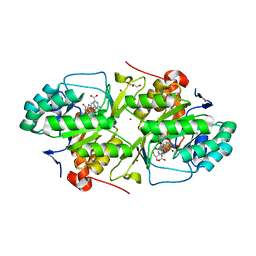 | | dihydroorotate dehydrogenase | | Descriptor: | BROMIDE ION, FLAVIN MONONUCLEOTIDE, GLYCEROL, ... | | Authors: | Arakaki, T.L, Merritt, E.A, Structural Genomics of Pathogenic Protozoa Consortium (SGPP) | | Deposit date: | 2005-09-23 | | Release date: | 2005-10-11 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Characterization of Trypanosoma brucei dihydroorotate dehydrogenase as a possible drug target; structural, kinetic and RNAi studies
Mol.Microbiol., 68, 2008
|
|
1JR0
 
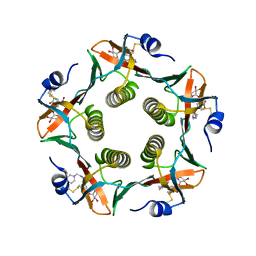 | | CHOLERA TOXIN B-PENTAMER WITH LIGAND BMSC-0011 | | Descriptor: | (3-NITRO-5-(2-MORPHOLIN-4-YL-ETHYLAMINOCARBONYL)PHENYL)-GALACTOPYRANOSIDE, cholera toxin B subunit | | Authors: | Merritt, E.A, Hol, W.G.J. | | Deposit date: | 2001-08-09 | | Release date: | 2002-05-08 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | Anchor-based design of improved cholera toxin and E. coli heat-labile enterotoxin receptor binding antagonists that display multiple binding modes.
Chem.Biol., 9, 2002
|
|
8B67
 
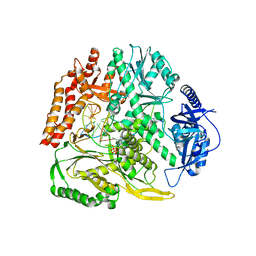 | |
8B7E
 
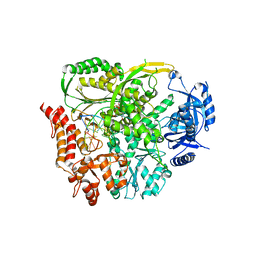 | |
8B77
 
 | |
8B6K
 
 | |
8B79
 
 | |
8B76
 
 | |
2X0N
 
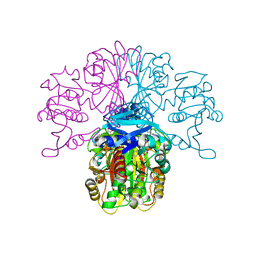 | | Structure of glycosomal glyceraldehyde-3-phosphate dehydrogenase from Trypanosoma brucei determined from Laue data | | Descriptor: | GLYCERALDEHYDE-3-PHOSPHATE DEHYDROGENASE, GLYCOSOMAL, NICOTINAMIDE-ADENINE-DINUCLEOTIDE, ... | | Authors: | Vellieux, F.M.D, Hajdu, J, Hol, W.G.J. | | Deposit date: | 2009-12-16 | | Release date: | 2009-12-22 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Structure of Glycosomal Glyceraldehyde-3-Phosphate Dehydrogenase from Trypanosoma Brucei Determined from Laue Data.
Proc.Natl.Acad.Sci.USA, 90, 1993
|
|
2TDX
 
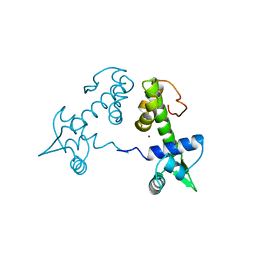 | | DIPHTHERIA TOX REPRESSOR (C102D MUTANT) COMPLEXED WITH NICKEL | | Descriptor: | DIPHTHERIA TOX REPRESSOR, NICKEL (II) ION | | Authors: | White, A, Ding, X, Zheng, H, Schiering, N, Ringe, D, Murphy, J.R. | | Deposit date: | 1998-06-22 | | Release date: | 1998-10-14 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structure of the metal-ion-activated diphtheria toxin repressor/tox operator complex.
Nature, 394, 1998
|
|
4MX9
 
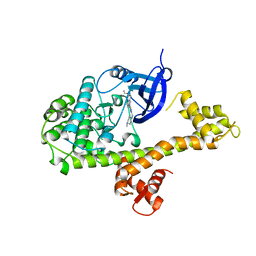 | | CDPK1 from Neospora caninum in complex with inhibitor UW1294 | | Descriptor: | 3-(6-ethoxynaphthalen-2-yl)-1-[(1-methylpiperidin-4-yl)methyl]-1H-pyrazolo[3,4-d]pyrimidin-4-amine, Calmodulin-like domain protein kinase isoenzyme gamma, related | | Authors: | Merritt, E.A. | | Deposit date: | 2013-09-26 | | Release date: | 2013-10-09 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Neospora caninum Calcium-Dependent Protein Kinase 1 Is an Effective Drug Target for Neosporosis Therapy.
Plos One, 9, 2014
|
|
4MXA
 
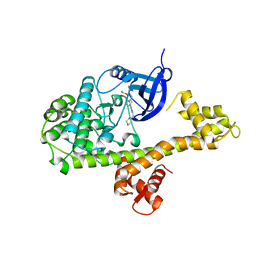 | | CDPK1 from Neospora caninum in complex with inhibitor RM-1-132 | | Descriptor: | 3-(6-ethoxynaphthalen-2-yl)-1-(piperidin-4-ylmethyl)-1H-pyrazolo[3,4-d]pyrimidin-4-amine, Calmodulin-like domain protein kinase isoenzyme gamma, related | | Authors: | Merritt, E.A. | | Deposit date: | 2013-09-26 | | Release date: | 2013-10-09 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Neospora caninum Calcium-Dependent Protein Kinase 1 Is an Effective Drug Target for Neosporosis Therapy.
Plos One, 9, 2014
|
|
1BI2
 
 | | STRUCTURE OF APO-AND HOLO-DIPHTHERIA TOXIN REPRESSOR | | Descriptor: | DIPHTHERIA TOXIN REPRESSOR | | Authors: | Pohl, E, Hol, W.G.J. | | Deposit date: | 1998-06-21 | | Release date: | 1999-06-22 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Motion of the DNA-binding domain with respect to the core of the diphtheria toxin repressor (DtxR) revealed in the crystal structures of apo- and holo-DtxR.
J.Biol.Chem., 273, 1998
|
|
1A7K
 
 | | GLYCOSOMAL GLYCERALDEHYDE-3-PHOSPHATE DEHYDROGENASE IN A MONOCLINIC CRYSTAL FORM | | Descriptor: | GLYCERALDEHYDE-3-PHOSPHATE DEHYDROGENASE, NICOTINAMIDE-ADENINE-DINUCLEOTIDE, PHOSPHATE ION | | Authors: | Kim, H, Hol, W.G.J. | | Deposit date: | 1998-03-16 | | Release date: | 1998-06-17 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Crystal structure of Leishmania mexicana glycosomal glyceraldehyde-3-phosphate dehydrogenase in a new crystal form confirms the putative physiological active site structure.
J.Mol.Biol., 278, 1998
|
|
1BI0
 
 | | STRUCTURE OF APO-AND HOLO-DIPHTHERIA TOXIN REPRESSOR | | Descriptor: | DIPHTHERIA TOXIN REPRESSOR, SULFATE ION, ZINC ION | | Authors: | Pohl, E, Hol, W.G. | | Deposit date: | 1998-06-21 | | Release date: | 1999-07-22 | | Last modified: | 2022-12-21 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Motion of the DNA-binding domain with respect to the core of the diphtheria toxin repressor (DtxR) revealed in the crystal structures of apo- and holo-DtxR.
J.Biol.Chem., 273, 1998
|
|
1BI1
 
 | | STRUCTURE OF APO-AND HOLO-DIPHTHERIA TOXIN REPRESSOR | | Descriptor: | DIPHTHERIA TOXIN REPRESSOR | | Authors: | Pohl, E, Hol, W.G.J. | | Deposit date: | 1998-06-21 | | Release date: | 1999-06-22 | | Last modified: | 2022-12-21 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Motion of the DNA-binding domain with respect to the core of the diphtheria toxin repressor (DtxR) revealed in the crystal structures of apo- and holo-DtxR.
J.Biol.Chem., 273, 1998
|
|
