7EQ2
 
 | | Crystal structure of GDP-bound Rab1a-T75D | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, CACODYLATE ION, COBALT HEXAMMINE(III), ... | | Authors: | Cao, Y.L, Gu, D.D, Gao, S. | | Deposit date: | 2021-04-28 | | Release date: | 2022-11-02 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.55090284 Å) | | Cite: | Aurora kinase A-mediated phosphorylation triggers structural alteration of Rab1A to enhance ER complexity during mitosis
Nat.Struct.Mol.Biol., 2024
|
|
7FCV
 
 | | Cryo-EM structure of the Potassium channel AKT1 mutant from Arabidopsis thaliana | | Descriptor: | PHOSPHATIDYLETHANOLAMINE, POTASSIUM ION, Potassium channel AKT1 | | Authors: | Yang, G.H, Lu, Y.M, Jia, Y.T, Zhang, Y.M, Tang, R.F, Xu, X, Li, X.M, Lei, J.L. | | Deposit date: | 2021-07-15 | | Release date: | 2022-11-09 | | Last modified: | 2024-06-12 | | Method: | ELECTRON MICROSCOPY (2.9 Å) | | Cite: | Structural basis for the activity regulation of a potassium channel AKT1 from Arabidopsis.
Nat Commun, 13, 2022
|
|
6LID
 
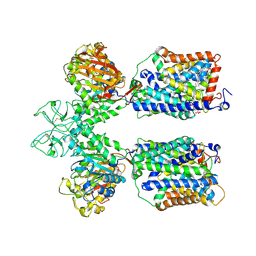 | | Heteromeric amino acid transporter b0,+AT-rBAT complex | | Descriptor: | 1,2-DIACYL-GLYCEROL-3-SN-PHOSPHATE, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, ... | | Authors: | Yan, R.H, Li, Y.N, Lei, J.L, Zhou, Q. | | Deposit date: | 2019-12-10 | | Release date: | 2020-04-29 | | Last modified: | 2024-11-06 | | Method: | ELECTRON MICROSCOPY (2.7 Å) | | Cite: | Cryo-EM structure of the human heteromeric amino acid transporter b0,+AT-rBAT.
Sci Adv, 6, 2020
|
|
6LI9
 
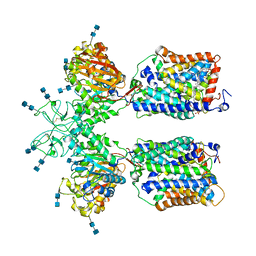 | | Heteromeric amino acid transporter b0,+AT-rBAT complex bound with Arginine | | Descriptor: | 1,2-DIACYL-GLYCEROL-3-SN-PHOSPHATE, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ARGININE, ... | | Authors: | Yan, R.H, Li, Y.N, Lei, J.L, Zhou, Q. | | Deposit date: | 2019-12-10 | | Release date: | 2020-04-29 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (2.3 Å) | | Cite: | Cryo-EM structure of the human heteromeric amino acid transporter b0,+AT-rBAT.
Sci Adv, 6, 2020
|
|
5XSY
 
 | | Structure of the Nav1.4-beta1 complex from electric eel | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Sodium channel protein, Voltage-gated sodium channel beta subunit 1, ... | | Authors: | Yan, Z, Zhou, Q, Wu, J.P, Yan, N. | | Deposit date: | 2017-06-15 | | Release date: | 2017-08-09 | | Last modified: | 2025-07-02 | | Method: | ELECTRON MICROSCOPY (4 Å) | | Cite: | Structure of the Nav1.4-beta 1 Complex from Electric Eel.
Cell, 170, 2017
|
|
9L4A
 
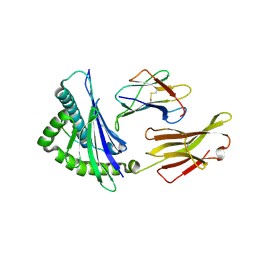 | | Crystal structure of HLA-C*12:02-MY9 | | Descriptor: | Beta-2-microglobulin, MHC class I antigen, MY9 | | Authors: | Yang, M, Zhong, P.L, Wei, P.C. | | Deposit date: | 2024-12-20 | | Release date: | 2025-03-19 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Biochemical and structural insights into position 97 micropolymorphisms in human leukocyte antigen (HLA)-C*12 allotypes and their differential disease associations.
Int.J.Biol.Macromol., 306, 2025
|
|
9L49
 
 | | Crystal structure of HLA-C*12:03-RV9 | | Descriptor: | Beta-2-microglobulin, MHC class I antigen, Toll-like receptor 9 | | Authors: | Yang, M, Zhong, P.L, Wei, P.C. | | Deposit date: | 2024-12-20 | | Release date: | 2025-03-19 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Biochemical and structural insights into position 97 micropolymorphisms in human leukocyte antigen (HLA)-C*12 allotypes and their differential disease associations.
Int.J.Biol.Macromol., 306, 2025
|
|
9L47
 
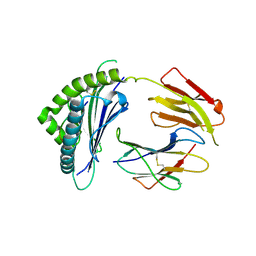 | | Crystal structure of HLA-C*12:03-MY9 | | Descriptor: | Beta-2-microglobulin, MHC class I antigen, MY9 | | Authors: | Yang, M, Zhong, P.L, Wei, P.C. | | Deposit date: | 2024-12-20 | | Release date: | 2025-03-19 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Biochemical and structural insights into position 97 micropolymorphisms in human leukocyte antigen (HLA)-C*12 allotypes and their differential disease associations.
Int.J.Biol.Macromol., 306, 2025
|
|
9L48
 
 | | Crystal structure of HLA-C*12:02-RV9 | | Descriptor: | Beta-2-microglobulin, MHC class I antigen, Toll-like receptor 9 | | Authors: | Yang, M, Zhong, P.L, Wei, P.C. | | Deposit date: | 2024-12-20 | | Release date: | 2025-03-19 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Biochemical and structural insights into position 97 micropolymorphisms in human leukocyte antigen (HLA)-C*12 allotypes and their differential disease associations.
Int.J.Biol.Macromol., 306, 2025
|
|
5XJY
 
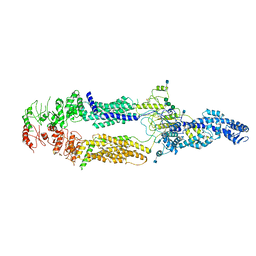 | | Cryo-EM structure of human ABCA1 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ATP-binding cassette sub-family A member 1, ... | | Authors: | Qian, H.W, Yan, N, Gong, X. | | Deposit date: | 2017-05-04 | | Release date: | 2017-06-07 | | Last modified: | 2025-07-02 | | Method: | ELECTRON MICROSCOPY (4.1 Å) | | Cite: | Structure of the Human Lipid Exporter ABCA1.
Cell, 169, 2017
|
|
7FIK
 
 | | The cryo-EM structure of the CR subunit from X. laevis NPC | | Descriptor: | MGC154553 protein, MGC83295 protein, MGC83926 protein, ... | | Authors: | Shi, Y, Huang, G, Zhan, X. | | Deposit date: | 2021-07-31 | | Release date: | 2022-11-09 | | Last modified: | 2024-06-12 | | Method: | ELECTRON MICROSCOPY (3.7 Å) | | Cite: | Structure of the cytoplasmic ring of the Xenopus laevis nuclear pore complex.
Science, 376, 2022
|
|
7ROQ
 
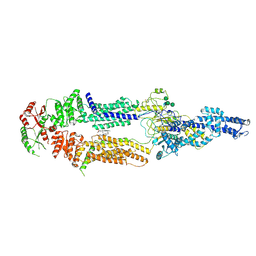 | | Alternative Structure of Human ABCA1 | | Descriptor: | (2S)-3-(hexadecanoyloxy)-2-[(9Z)-octadec-9-enoyloxy]propyl 2-(trimethylammonio)ethyl phosphate, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Aller, S.G. | | Deposit date: | 2021-08-01 | | Release date: | 2022-01-19 | | Last modified: | 2025-05-28 | | Method: | ELECTRON MICROSCOPY (4.1 Å) | | Cite: | Refined ABCA1 with Domain-swapped Regulatory Domains
Plos One, 2022
|
|
7FIL
 
 | |
3EP2
 
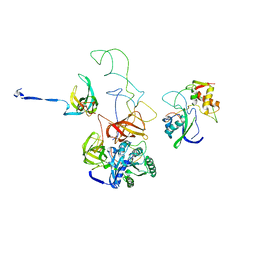 | | Model of Phe-tRNA(Phe) in the ribosomal pre-accommodated state revealed by cryo-EM | | Descriptor: | 30S ribosomal protein S12, 50S ribosomal protein L11, Elongation factor Tu, ... | | Authors: | Frank, J, Li, W, Agirrezabala, X. | | Deposit date: | 2008-09-29 | | Release date: | 2008-12-16 | | Last modified: | 2024-02-21 | | Method: | ELECTRON MICROSCOPY (9 Å) | | Cite: | Recognition of aminoacyl-tRNA: a common molecular mechanism revealed by cryo-EM.
Embo J., 27, 2008
|
|
3EQ4
 
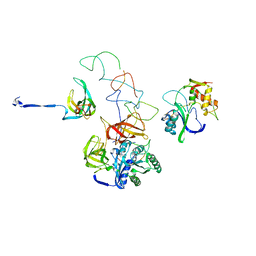 | | Model of tRNA(Leu)-EF-Tu in the ribosomal pre-accommodated state revealed by cryo-EM | | Descriptor: | 30S ribosomal protein S12, 50S ribosomal protein L11, Elongation factor Tu, ... | | Authors: | Frank, J, Li, W, Agirrezabala, X. | | Deposit date: | 2008-09-30 | | Release date: | 2008-12-16 | | Last modified: | 2024-02-21 | | Method: | ELECTRON MICROSCOPY (12 Å) | | Cite: | Recognition of aminoacyl-tRNA: a common molecular mechanism revealed by cryo-EM.
Embo J., 27, 2008
|
|
3EQ3
 
 | | Model of tRNA(Trp)-EF-Tu in the ribosomal pre-accommodated state revealed by cryo-EM | | Descriptor: | 30S ribosomal protein S12, 50S ribosomal protein L11, Elongation factor Tu, ... | | Authors: | Frank, J, Li, W, Agirrezabala, X. | | Deposit date: | 2008-09-30 | | Release date: | 2008-12-16 | | Last modified: | 2024-02-21 | | Method: | ELECTRON MICROSCOPY (9 Å) | | Cite: | Recognition of aminoacyl-tRNA: a common molecular mechanism revealed by cryo-EM.
Embo J., 27, 2008
|
|
6KU9
 
 | |
6L9N
 
 | | H2-Ld complexed with A5 peptide | | Descriptor: | MHC, SER-PRO-SER-TYR-ALA-TYR-HIS-GLN-PHE, b2m | | Authors: | Wei, P.C, Yin, L. | | Deposit date: | 2019-11-10 | | Release date: | 2020-11-18 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Structures suggest an approach for converting weak self-peptide tumor antigens into superagonists for CD8 T cells in cancer.
Proc.Natl.Acad.Sci.USA, 118, 2021
|
|
6KZP
 
 | | calcium channel-ligand | | Descriptor: | 1,2-Distearoyl-sn-glycerophosphoethanolamine, 2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, ... | | Authors: | Yan, N. | | Deposit date: | 2019-09-25 | | Release date: | 2019-12-18 | | Last modified: | 2024-11-06 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Cryo-EM structures of apo and antagonist-bound human Cav3.1.
Nature, 576, 2019
|
|
6L9L
 
 | | 1D4 TCR recognition of H2-Ld a1a2 A5 Peptide Complexes | | Descriptor: | H2-Ld a1a2, SER-PRO-SER-TYR-ALA-TYR-HIS-GLN-PHE, T Cell Receptor | | Authors: | Wei, P.C, Yin, L. | | Deposit date: | 2019-11-10 | | Release date: | 2020-11-18 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.399 Å) | | Cite: | Structures suggest an approach for converting weak self-peptide tumor antigens into superagonists for CD8 T cells in cancer.
Proc.Natl.Acad.Sci.USA, 118, 2021
|
|
6L9M
 
 | | H2-Ld complexed with AH1 peptide | | Descriptor: | H2-Ld, SER-PRO-SER-TYR-VAL-TYR-HIS-GLN-PHE, b2m | | Authors: | Wei, P.C, Yin, L. | | Deposit date: | 2019-11-10 | | Release date: | 2020-11-18 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Structures suggest an approach for converting weak self-peptide tumor antigens into superagonists for CD8 T cells in cancer.
Proc.Natl.Acad.Sci.USA, 118, 2021
|
|
6KZO
 
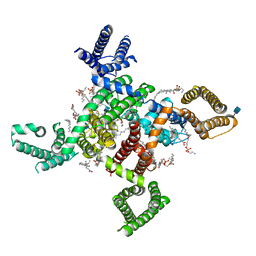 | | membrane protein | | Descriptor: | 1,2-Distearoyl-sn-glycerophosphoethanolamine, 2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, ... | | Authors: | Yan, N. | | Deposit date: | 2019-09-25 | | Release date: | 2019-12-18 | | Last modified: | 2024-10-30 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Cryo-EM structures of apo and antagonist-bound human Cav3.1.
Nature, 576, 2019
|
|
6LK8
 
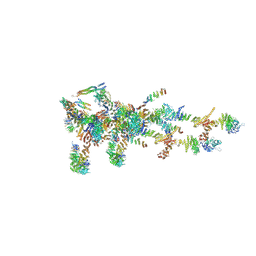 | | Structure of Xenopus laevis Cytoplasmic Ring subunit. | | Descriptor: | GATOR complex protein SEC13, MGC154553 protein, MGC83295 protein, ... | | Authors: | Shi, Y, Huang, G, Yan, C, Zhang, Y. | | Deposit date: | 2019-12-18 | | Release date: | 2021-07-21 | | Last modified: | 2024-05-29 | | Method: | ELECTRON MICROSCOPY (5.5 Å) | | Cite: | Structure of the cytoplasmic ring of the Xenopus laevis nuclear pore complex by cryo-electron microscopy single particle analysis.
Cell Res., 30, 2020
|
|
8X94
 
 | | Structure of human TRPV1 in complex with antagonist --protein purified without CHS | | Descriptor: | 4-(7-Hydroxy-2-isopropyl-4-oxoquinazolin-3(4H)-yl)benzonitrile, CHOLESTEROL, Transient receptor potential cation channel subfamily V member 1,Green fluorescent protein | | Authors: | Fan, J, Lei, X. | | Deposit date: | 2023-11-29 | | Release date: | 2024-08-14 | | Last modified: | 2024-08-21 | | Method: | ELECTRON MICROSCOPY (2.55 Å) | | Cite: | Structural basis of TRPV1 inhibition by SAF312 and cholesterol.
Nat Commun, 15, 2024
|
|
8JQR
 
 | | Structure of human TRPV1 in complex with antagonist | | Descriptor: | 4-(7-Hydroxy-2-isopropyl-4-oxoquinazolin-3(4H)-yl)benzonitrile, CHOLESTEROL, Transient receptor potential cation channel subfamily V member 1,PreScission Site,Green fluorescent protein | | Authors: | Fan, J, Lei, X. | | Deposit date: | 2023-06-14 | | Release date: | 2024-08-14 | | Last modified: | 2024-08-21 | | Method: | ELECTRON MICROSCOPY (2.75 Å) | | Cite: | Structural basis of TRPV1 inhibition by SAF312 and cholesterol.
Nat Commun, 15, 2024
|
|
