2OR3
 
 | |
2OQ0
 
 | | Crystal Structure of the First HIN-200 Domain of Interferon-Inducible Protein 16 | | Descriptor: | CHLORIDE ION, Gamma-interferon-inducible protein Ifi-16 | | Authors: | Lam, R, Liao, J.C.C, Ravichandran, M, Ma, J, Tempel, W, Chirgadze, N.Y, Arrowsmith, C.H, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2007-01-30 | | Release date: | 2007-02-27 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal Structure of the First HIN-200 Domain of Interferon-Inducible Protein 16
To be Published
|
|
6QNV
 
 | | Fibrinogen-like globe domain of Human Tenascin-C | | Descriptor: | Tenascin | | Authors: | Coker, J.A, Bezerra, G.A, Bradshaw, W.J, Zhang, M, Yosaatmadja, Y, Fernandez-Cid, A, Shrestha, L, Burgess-Brown, N, Gileadi, O, Arrowsmith, C.H, Bountra, C, Midwood, K.S, Yue, W.W, Marsden, B.D, Structural Genomics Consortium (SGC) | | Deposit date: | 2019-02-12 | | Release date: | 2019-02-27 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Fibrinogen-like globe domain of Human Tenascin-C
To Be Published
|
|
1U38
 
 | | Auto-inhibition Mechanism of X11s/Mints Family Scaffold Proteins Revealed by the Closed Conformation of the Tandem PDZ Domains | | Descriptor: | PVYI, amyloid beta A4 precursor protein-binding, family A, ... | | Authors: | Feng, W, Long, J.-F, Chan, L.-N, He, C, Fu, A, Xia, J, Ip, N.Y, Zhang, M. | | Deposit date: | 2004-07-21 | | Release date: | 2005-07-26 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Autoinhibition of X11/Mint scaffold proteins revealed by the closed conformation of the PDZ tandem
Nat.Struct.Mol.Biol., 12, 2005
|
|
1U3B
 
 | | Auto-inhibition Mechanism of X11s/Mints Family Scaffold Proteins Revealed by the Closed Conformation of the Tandem PDZ Domains | | Descriptor: | amyloid beta A4 precursor protein-binding, family A, member 1 | | Authors: | Feng, W, Long, J.-F, Chan, L.-N, He, C, Fu, A, Xia, J, Ip, N.Y, Zhang, M. | | Deposit date: | 2004-07-21 | | Release date: | 2005-07-26 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Autoinhibition of X11/Mint scaffold proteins revealed by the closed conformation of the PDZ tandem
Nat.Struct.Mol.Biol., 12, 2005
|
|
1U37
 
 | | Auto-inhibition Mechanism of X11s/Mints Family Scaffold Proteins Revealed by the Closed Conformation of the Tandem PDZ Domains | | Descriptor: | amyloid beta A4 precursor protein-binding, family A, member 1 | | Authors: | Feng, W, Long, J.-F, Chan, L.-N, He, C, Fu, A, Xia, J, Ip, N.Y, Zhang, M. | | Deposit date: | 2004-07-21 | | Release date: | 2005-07-26 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Autoinhibition of X11/Mint scaffold proteins revealed by the closed conformation of the PDZ tandem
Nat.Struct.Mol.Biol., 12, 2005
|
|
1U39
 
 | | Auto-inhibition Mechanism of X11s/Mints Family Scaffold Proteins Revealed by the Closed Conformation of the Tandem PDZ Domains | | Descriptor: | amyloid beta A4 precursor protein-binding, family A, member 1 | | Authors: | Feng, W, Long, J.-F, Chan, L.-N, He, C, Fu, A, Xia, J, Ip, N.Y, Zhang, M. | | Deposit date: | 2004-07-21 | | Release date: | 2005-07-26 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Autoinhibition of X11/Mint scaffold proteins revealed by the closed conformation of the PDZ tandem
Nat.Struct.Mol.Biol., 12, 2005
|
|
5VBL
 
 | | Structure of apelin receptor in complex with agonist peptide | | Descriptor: | (2R)-2,3-dihydroxypropyl (9Z)-octadec-9-enoate, Apelin receptor,Rubredoxin,Apelin receptor Chimera, ZINC ION, ... | | Authors: | Ma, Y, Yue, Y, Ma, Y, Zhang, Q, Zhou, Q, Song, Y, Shen, Y, Li, X, Ma, X, Li, C, Hanson, M.A, Han, G.W, Sickmier, E.A, Swaminath, G, Zhao, S, Stevems, R.C, Hu, L.A, Zhong, W, Zhang, M, Xu, F. | | Deposit date: | 2017-03-29 | | Release date: | 2017-05-31 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Structural Basis for Apelin Control of the Human Apelin Receptor
Structure, 25, 2017
|
|
7SWL
 
 | | CryoEM structure of the N-terminal-deleted Rix7 AAA-ATPase | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, ADENOSINE-5'-TRIPHOSPHATE, MAGNESIUM ION, ... | | Authors: | Kocaman, S, Stanley, R.E, Lo, Y.H, Krahn, J, Dandey, V.P, Sobhany, M, Petrovich, M, Williams, J.G, Deterding, L.J, Borgnia, M.J, Etigunta, S. | | Deposit date: | 2021-11-20 | | Release date: | 2022-03-02 | | Last modified: | 2024-06-05 | | Method: | ELECTRON MICROSCOPY (2.88 Å) | | Cite: | Communication network within the essential AAA-ATPase Rix7 drives ribosome assembly.
Pnas Nexus, 1, 2022
|
|
7T0V
 
 | | CryoEM structure of the crosslinked Rix7 AAA-ATPase | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, ADENOSINE-5'-TRIPHOSPHATE, MAGNESIUM ION, ... | | Authors: | Kocaman, S, Stanley, R.E, Lo, Y.H, Krahn, J, Dandey, V.P, Sobhany, M, Petrovich, M, Williams, J.G, Deterding, L.J, Borgnia, M.J, Etigunta, S. | | Deposit date: | 2021-11-30 | | Release date: | 2022-03-02 | | Last modified: | 2024-06-05 | | Method: | ELECTRON MICROSCOPY (3.67 Å) | | Cite: | Communication network within the essential AAA-ATPase Rix7 drives ribosome assembly.
Pnas Nexus, 1, 2022
|
|
6LAD
 
 | |
5XBF
 
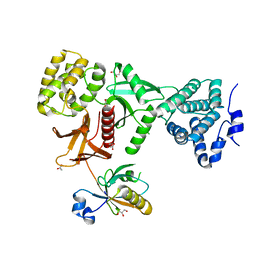 | | Crystal Structure of Myo7b C-terminal MyTH4-FERM in complex with USH1C PDZ3 | | Descriptor: | ACETATE ION, D-MALATE, GLYCEROL, ... | | Authors: | Li, J, He, Y, Weck, W.L, Lu, Q, Tyska, M.J, Zhang, M. | | Deposit date: | 2017-03-17 | | Release date: | 2017-05-17 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (1.802 Å) | | Cite: | Structure of Myo7b/USH1C complex suggests a general PDZ domain binding mode by MyTH4-FERM myosins.
Proc. Natl. Acad. Sci. U.S.A., 114, 2017
|
|
6KHX
 
 | | Crystal structure of Prx from Akkermansia muciniphila | | Descriptor: | CALCIUM ION, Peroxiredoxin | | Authors: | Li, M, Wang, J, Xu, W, Wang, Y, Zhang, M, Wang, M. | | Deposit date: | 2019-07-16 | | Release date: | 2020-02-19 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.58 Å) | | Cite: | Crystal structure of Akkermansia muciniphila peroxiredoxin reveals a novel regulatory mechanism of typical 2-Cys Prxs by a distinct loop.
Febs Lett., 594, 2020
|
|
6KY4
 
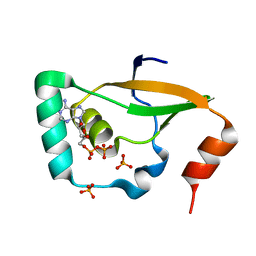 | | Crystal structure of Sulfiredoxin from Arabidopsis thaliana | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, PHOSPHATE ION, Sulfiredoxin, ... | | Authors: | Liu, M, Wang, J, Li, X, Li, M, Sylvanno, M.J, Zhang, M, Wang, M. | | Deposit date: | 2019-09-16 | | Release date: | 2019-10-16 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | The crystal structure of sulfiredoxin from Arabidopsis thaliana revealed a more robust antioxidant mechanism in plants.
Biochem.Biophys.Res.Commun., 520, 2019
|
|
6KYH
 
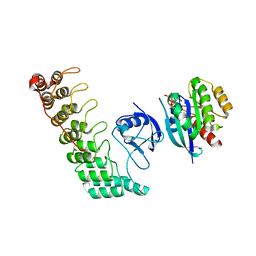 | | Crystal structure of Shank3 NTD-ANK A42K mutant in complex with HRas | | Descriptor: | GTPase HRas, MAGNESIUM ION, PHOSPHOAMINOPHOSPHONIC ACID-GUANYLATE ESTER, ... | | Authors: | Cai, Q, Zhang, M. | | Deposit date: | 2019-09-18 | | Release date: | 2019-12-04 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (3.3 Å) | | Cite: | Shank3 Binds to and Stabilizes the Active Form of Rap1 and HRas GTPases via Its NTD-ANK Tandem with Distinct Mechanisms.
Structure, 28, 2020
|
|
5B8D
 
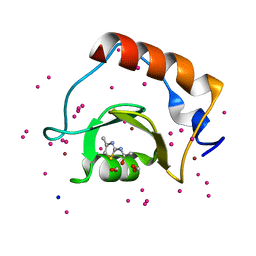 | | Crystal structure of a low occupancy fragment candidate (N-(4-Methyl-1,3-thiazol-2-yl)propanamide) bound adjacent to the ubiquitin binding pocket of the HDAC6 zinc-finger domain | | Descriptor: | FORMIC ACID, Histone deacetylase 6, SODIUM ION, ... | | Authors: | Harding, R.J, Tempel, W, Collins, P, Pearce, N, Brandao-Neto, J, Douangamath, A, Ravichandran, M, Schapira, M, Bountra, C, Edwards, A.M, von Delft, F, Santhakumar, V, Arrowsmith, C.M, Structural Genomics Consortium (SGC) | | Deposit date: | 2016-06-14 | | Release date: | 2016-07-27 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.05 Å) | | Cite: | Small Molecule Antagonists of the Interaction between the Histone Deacetylase 6 Zinc-Finger Domain and Ubiquitin.
J. Med. Chem., 60, 2017
|
|
6KYK
 
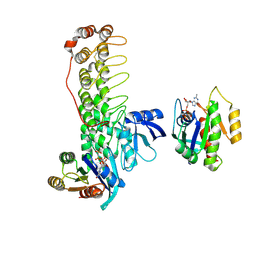 | | Crystal structure of Shank3 NTD-ANK mutant in complex with Rap1 | | Descriptor: | MAGNESIUM ION, PHOSPHOAMINOPHOSPHONIC ACID-GUANYLATE ESTER, Ras-related protein Rap-1b, ... | | Authors: | Cai, Q, Zhang, M. | | Deposit date: | 2019-09-19 | | Release date: | 2019-12-04 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.82 Å) | | Cite: | Shank3 Binds to and Stabilizes the Active Form of Rap1 and HRas GTPases via Its NTD-ANK Tandem with Distinct Mechanisms.
Structure, 28, 2020
|
|
6KZJ
 
 | | Crystal structure of Ankyrin B/NdeL1 complex | | Descriptor: | Ankyrin-2, Nuclear distribution protein nudE-like 1 | | Authors: | Ye, J, Li, J, Ye, F, Zhang, M, Zhang, Y, Wang, C. | | Deposit date: | 2019-09-24 | | Release date: | 2020-01-15 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Mechanistic insights into the interactions of dynein regulator Ndel1 with neuronal ankyrins and implications in polarity maintenance.
Proc.Natl.Acad.Sci.USA, 117, 2020
|
|
1GGZ
 
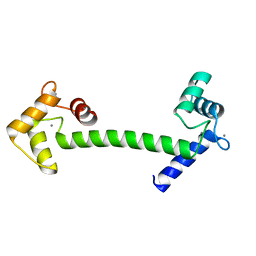 | | CRYSTAL STRUCTURE OF THE CALMODULIN-LIKE PROTEIN (HCLP) FROM HUMAN EPITHELIAL CELLS | | Descriptor: | CALCIUM ION, CALMODULIN-RELATED PROTEIN NB-1 | | Authors: | Han, B.-G, Han, M, Sui, H, Yaswen, P, Walian, P.J, Jap, B.K. | | Deposit date: | 2000-10-13 | | Release date: | 2002-06-05 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Crystal structure of human calmodulin-like protein: insights into its functional role.
FEBS Lett., 521, 2002
|
|
5C1T
 
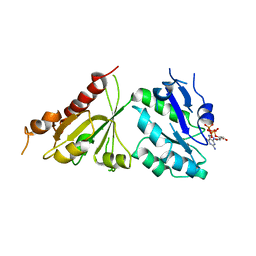 | | Crystal structure of the GTP-bound wild type EhRabX3 from Entamoeba histolytica | | Descriptor: | GUANOSINE-5'-TRIPHOSPHATE, MAGNESIUM ION, Small GTPase EhRabX3 | | Authors: | Srivastava, V.K, Chandra, M, Datta, S. | | Deposit date: | 2015-06-15 | | Release date: | 2016-04-27 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.801 Å) | | Cite: | Crystal Structure Analysis of Wild Type and Fast Hydrolyzing Mutant of EhRabX3, a Tandem Ras Superfamily GTPase from Entamoeba histolytica.
J.Mol.Biol., 428, 2016
|
|
8OWJ
 
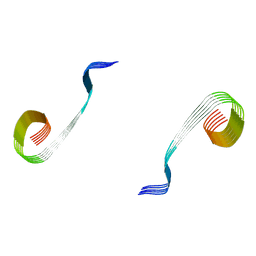 | | Lipidic amyloid-beta(1-40) fibril - polymorph L2-L2 | | Descriptor: | Amyloid-beta A4 protein | | Authors: | Frieg, B, Han, M, Giller, K, Dienemann, C, Riedel, D, Becker, S, Andreas, L.B, Griesinger, C, Schroeder, G.F. | | Deposit date: | 2023-04-28 | | Release date: | 2024-03-06 | | Method: | ELECTRON MICROSCOPY (3.75 Å) | | Cite: | Cryo-EM structures of lipidic fibrils of amyloid-beta (1-40).
Nat Commun, 15, 2024
|
|
8OVM
 
 | | Lipidic amyloid-beta(1-40) fibril - polymorph L2 | | Descriptor: | Amyloid-beta A4 protein | | Authors: | Frieg, B, Han, M, Giller, K, Dienemann, C, Riedel, D, Becker, S, Andreas, L.B, Griesinger, C, Schroeder, G.F. | | Deposit date: | 2023-04-26 | | Release date: | 2024-03-06 | | Method: | ELECTRON MICROSCOPY (3.24 Å) | | Cite: | Cryo-EM structures of lipidic fibrils of amyloid-beta (1-40).
Nat Commun, 15, 2024
|
|
8OWE
 
 | | Lipidic amyloid-beta(1-40) fibril - polymorph L2-L3 | | Descriptor: | Amyloid-beta A4 protein | | Authors: | Frieg, B, Han, M, Giller, K, Dienemann, C, Riedel, D, Becker, S, Andreas, L.B, Griesinger, C, Schroeder, G.F. | | Deposit date: | 2023-04-27 | | Release date: | 2024-03-06 | | Method: | ELECTRON MICROSCOPY (3.75 Å) | | Cite: | Cryo-EM structures of lipidic fibrils of amyloid-beta (1-40).
Nat Commun, 15, 2024
|
|
8OVK
 
 | | Lipidic amyloid-beta(1-40) fibril - polymorph L1 | | Descriptor: | Amyloid-beta A4 protein | | Authors: | Frieg, B, Han, M, Giller, K, Dienemann, C, Riedel, D, Becker, S, Andreas, L.B, Griesinger, C, Schroeder, G.F. | | Deposit date: | 2023-04-26 | | Release date: | 2024-03-06 | | Method: | ELECTRON MICROSCOPY (2.88 Å) | | Cite: | Cryo-EM structures of lipidic fibrils of amyloid-beta (1-40).
Nat Commun, 15, 2024
|
|
8OWK
 
 | | Lipidic amyloid-beta(1-40) fibril - polymorph L3-L3 | | Descriptor: | Amyloid-beta A4 protein | | Authors: | Frieg, B, Han, M, Giller, K, Dienemann, C, Riedel, D, Becker, S, Andreas, L.B, Griesinger, C, Schroeder, G.F. | | Deposit date: | 2023-04-28 | | Release date: | 2024-03-06 | | Method: | ELECTRON MICROSCOPY (3.86 Å) | | Cite: | Cryo-EM structures of lipidic fibrils of amyloid-beta (1-40).
Nat Commun, 15, 2024
|
|
