1PBD
 
 | | CRYSTAL STRUCTURES OF WILD-TYPE P-HYDROXYBENZOATE HYDROXYLASE COMPLEXED WITH 4-AMINOBENZOATE, 2,4-DIHYDROXYBENZOATE AND 2-HYDROXY-4-AMINOBENZOATE AND OF THE TRY222ALA MUTANT, COMPLEXED WITH 2-HYDROXY-4-AMINOBENZOATE. EVIDENCE FOR A PROTON CHANNEL AND A NEW BINDING MODE OF THE FLAVIN RING | | Descriptor: | 4-AMINOBENZOIC ACID, FLAVIN-ADENINE DINUCLEOTIDE, P-HYDROXYBENZOATE HYDROXYLASE | | Authors: | Schreuder, H.A, Mattevi, A, Hol, W.G.J. | | Deposit date: | 1994-07-06 | | Release date: | 1994-09-30 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystal structures of wild-type p-hydroxybenzoate hydroxylase complexed with 4-aminobenzoate,2,4-dihydroxybenzoate, and 2-hydroxy-4-aminobenzoate and of the Tyr222Ala mutant complexed with 2-hydroxy-4-aminobenzoate. Evidence for a proton channel and a new binding mode of the flavin ring
Biochemistry, 33, 1994
|
|
4B64
 
 | | A. fumigatus ornithine hydroxylase (SidA) bound to NADP and Lysine | | Descriptor: | FLAVIN-ADENINE DINUCLEOTIDE, GLYCEROL, L-ORNITHINE N5 MONOOXYGENASE, ... | | Authors: | Franceschini, S, Fedkenheuer, M, Vogelaar, N.J, Robinson, H.H, Sobrado, P, Mattevi, A. | | Deposit date: | 2012-08-09 | | Release date: | 2012-10-03 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.28 Å) | | Cite: | Structural Insight Into the Mechanism of Oxygen Activation and Substrate Selectivity of Flavin-Dependent N-Hydroxylating Monooxygenases.
Biochemistry, 51, 2012
|
|
1EA0
 
 | | Alpha subunit of A. brasilense glutamate synthase | | Descriptor: | 2-OXOGLUTARIC ACID, FE3-S4 CLUSTER, FLAVIN MONONUCLEOTIDE, ... | | Authors: | Binda, C, Bossi, R.T, Vanoni, M.A, Mattevi, A. | | Deposit date: | 2000-11-02 | | Release date: | 2001-11-01 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Cross-Talk and Ammonia Channeling between Active Centers in the Unexpected Domain Arrangement of Glutamate Synthase
Structure, 8, 2000
|
|
5L3C
 
 | | Human LSD1/CoREST: LSD1 E379K mutation | | Descriptor: | FLAVIN-ADENINE DINUCLEOTIDE, Lysine-specific histone demethylase 1A, REST corepressor 1 | | Authors: | Pilotto, S, Speranzini, V, Marabelli, C, Mattevi, A. | | Deposit date: | 2016-04-06 | | Release date: | 2016-05-04 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (3.31 Å) | | Cite: | LSD1/KDM1A mutations associated to a newly described form of intellectual disability impair demethylase activity and binding to transcription factors.
Hum.Mol.Genet., 25, 2016
|
|
5M0Z
 
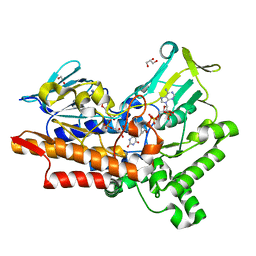 | |
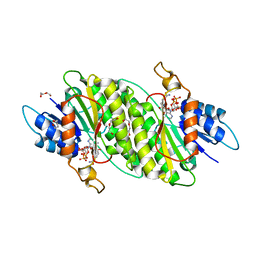
5MXU
 
 | | Structure of the Y503F mutant of vanillyl alcohol oxidase | | Descriptor: | FLAVIN-ADENINE DINUCLEOTIDE, GLYCEROL, Vanillyl-alcohol oxidase | | Authors: | Ewing, T.A, Nguyen, Q.-T, Allan, R.C, Gygli, G, Romero, E, Binda, C, Fraaije, M.W, Mattevi, A, van Berkel, W.J.H. | | Deposit date: | 2017-01-24 | | Release date: | 2017-07-26 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Two tyrosine residues, Tyr-108 and Tyr-503, are responsible for the deprotonation of phenolic substrates in vanillyl-alcohol oxidase.
J. Biol. Chem., 292, 2017
|
|
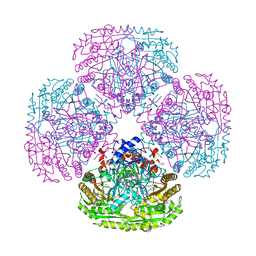
5MLN
 
 | |
5MXJ
 
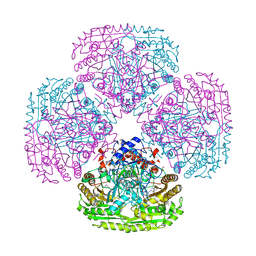 | | Structure of the Y108F mutant of vanillyl alcohol oxidase | | Descriptor: | FLAVIN-ADENINE DINUCLEOTIDE, GLYCEROL, Vanillyl-alcohol oxidase | | Authors: | Ewing, T.A, Nguyen, Q.-T, Allan, R.C, Gygli, G, Romero, E, Binda, C, Fraaije, M.W, Mattevi, A, van Berkel, W.J.H. | | Deposit date: | 2017-01-23 | | Release date: | 2017-07-26 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Two tyrosine residues, Tyr-108 and Tyr-503, are responsible for the deprotonation of phenolic substrates in vanillyl-alcohol oxidase.
J. Biol. Chem., 292, 2017
|
|
5MQ6
 
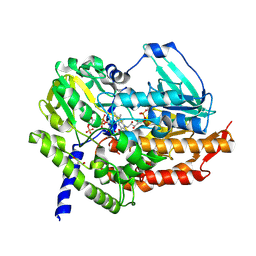 | | Polycyclic Ketone Monooxygenase from the Thermophilic Fungus Thermothelomyces thermophila | | Descriptor: | FLAVIN-ADENINE DINUCLEOTIDE, GLYCEROL, NADPH DIHYDRO-NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, ... | | Authors: | Savino, S, Furst, M.J.L.J, Fraaije, M.W, Mattevi, A. | | Deposit date: | 2016-12-20 | | Release date: | 2017-01-11 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Polycyclic Ketone Monooxygenase from the Thermophilic Fungus Thermothelomyces thermophila: A Structurally Distinct Biocatalyst for Bulky Substrates.
J. Am. Chem. Soc., 139, 2017
|
|
4AP3
 
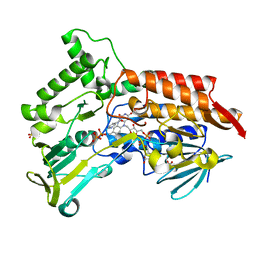 | | Oxidized steroid monooxygenase bound to NADP | | Descriptor: | FLAVIN-ADENINE DINUCLEOTIDE, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, STEROID MONOOXYGENASE, ... | | Authors: | Franceschini, S, van Beek, H.L, Martinoli, C, Pennetta, A, Fraaije, M.W, Mattevi, A. | | Deposit date: | 2012-03-30 | | Release date: | 2012-05-30 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.39 Å) | | Cite: | Exploring the Structural Basis of Substrate Preferences in Baeyer-Villiger Monooxygenases: Insight from Steroid Monooxygenase.
J.Biol.Chem., 287, 2012
|
|
5LXE
 
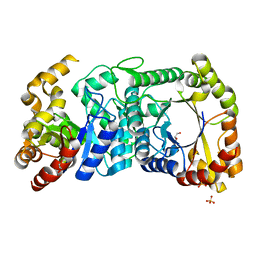 | | F420-dependent glucose-6-phosphate dehydrogenase from Rhodococcus jostii RHA1 | | Descriptor: | F420-dependent glucose-6-phosphate dehydrogenase 1, GLYCEROL, SULFATE ION | | Authors: | Nguyen, Q.-T, Trinco, G, Binda, C, Mattevi, A, Fraaije, M.W. | | Deposit date: | 2016-09-20 | | Release date: | 2016-12-28 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.47 Å) | | Cite: | Discovery and characterization of an F420-dependent glucose-6-phosphate dehydrogenase (Rh-FGD1) from Rhodococcus jostii RHA1.
Appl. Microbiol. Biotechnol., 101, 2017
|
|
5N2I
 
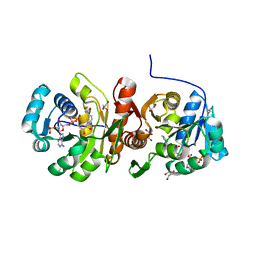 | | F420:NADPH oxidoreductase from Thermobifida fusca with NADP+ bound | | Descriptor: | GLYCEROL, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, Reduced coenzyme F420:NADP oxidoreductase | | Authors: | Kumar, H, Nguyen, Q.-T, Binda, C, Mattevi, A, Fraaije, M.W. | | Deposit date: | 2017-02-07 | | Release date: | 2017-04-26 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Isolation and characterization of a thermostable F420:NADPH oxidoreductase from Thermobifida fusca.
J. Biol. Chem., 292, 2017
|
|
5M10
 
 | |
4AP1
 
 | | Oxidized steroid monooxygenase bound to NADP | | Descriptor: | FLAVIN-ADENINE DINUCLEOTIDE, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, STEROID MONOOXYGENASE, ... | | Authors: | Franceschini, S, van Beek, H.L, Martinoli, C, Pennetta, A, Fraaije, M.W, Mattevi, A. | | Deposit date: | 2012-03-30 | | Release date: | 2012-05-30 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.95 Å) | | Cite: | Exploring the Structural Basis of Substrate Preferences in Baeyer-Villiger Monooxygenases: Insight from Steroid Monooxygenase.
J.Biol.Chem., 287, 2012
|
|
4BJZ
 
 | | Crystal structure of 3-hydroxybenzoate 6-hydroxylase uncovers lipid- assisted flavoprotein strategy for regioselective aromatic hydroxylation: Native data | | Descriptor: | CHLORIDE ION, FLAVIN-ADENINE DINUCLEOTIDE, PHOSPHATIDYLGLYCEROL-PHOSPHOGLYCEROL, ... | | Authors: | Orru, R, Montersino, S, Barendregt, A, Westphal, A.H, van Duijn, E, Mattevi, A, van Berkel, W.J.H. | | Deposit date: | 2013-04-21 | | Release date: | 2013-07-24 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.51 Å) | | Cite: | Crystal Structure of 3-Hydroxybenzoate 6-Hydroxylase Uncovers Lipid-Assisted Flavoprotein Strategy for Regioselective Aromatic Hydroxylation
J.Biol.Chem., 288, 2013
|
|
4BBY
 
 | | MAMMALIAN ALKYLDIHYDROXYACETONEPHOSPHATE SYNTHASE: WILD-TYPE | | Descriptor: | ALKYLDIHYDROXYACETONEPHOSPHATE SYNTHASE, PEROXISOMAL, FLAVIN-ADENINE DINUCLEOTIDE, ... | | Authors: | Nenci, S, Piano, V, Rosati, S, Aliverti, A, Pandini, V, Fraaije, M.W, Heck, A.J.R, Edmondson, D.E, Mattevi, A. | | Deposit date: | 2012-09-28 | | Release date: | 2012-11-07 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Precursor of Ether Phospholipids is Synthesized by a Flavoenzyme Through Covalent Catalysis.
Proc.Natl.Acad.Sci.USA, 109, 2012
|
|
4BJY
 
 | | Crystal structure of 3-hydroxybenzoate 6-hydroxylase uncovers lipid- assisted flavoprotein strategy for regioselective aromatic hydroxylation: Platinum derivative | | Descriptor: | CHLORIDE ION, FLAVIN-ADENINE DINUCLEOTIDE, PHOSPHATIDYLGLYCEROL-PHOSPHOGLYCEROL, ... | | Authors: | Orru, R, Montersino, S, Barendregt, A, Westphal, A.H, van Duijn, E, Mattevi, A, van Berkel, W.J.H. | | Deposit date: | 2013-04-21 | | Release date: | 2013-07-24 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.52 Å) | | Cite: | Crystal Structure of 3-Hydroxybenzoate 6-Hydroxylase Uncovers Lipid-Assisted Flavoprotein Strategy for Regioselective Aromatic Hydroxylation
J.Biol.Chem., 288, 2013
|
|
4BK1
 
 | | Crystal structure of 3-hydroxybenzoate 6-hydroxylase uncovers lipid- assisted flavoprotein strategy for regioselective aromatic hydroxylation: H213S mutant in complex with 3-hydroxybenzoate | | Descriptor: | 3-HYDROXYBENZOIC ACID, CHLORIDE ION, FLAVIN-ADENINE DINUCLEOTIDE, ... | | Authors: | Orru, R, Montersino, S, Barendregt, A, Westphal, A.H, van Duijn, E, Mattevi, A, van Berkel, W.J.H. | | Deposit date: | 2013-04-21 | | Release date: | 2013-07-24 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.73 Å) | | Cite: | Crystal Structure of 3-Hydroxybenzoate 6-Hydroxylase Uncovers Lipid-Assisted Flavoprotein Strategy for Regioselective Aromatic Hydroxylation
J.Biol.Chem., 288, 2013
|
|
4BCA
 
 | | MAMMALIAN ALKYLDIHYDROXYACETONEPHOSPHATE SYNTHASE: Tyr578Phe mutant | | Descriptor: | ALKYLDIHYDROXYACETONEPHOSPHATE SYNTHASE, PEROXISOMAL, CHLORIDE ION, ... | | Authors: | Nenci, S, Piano, V, Rosati, S, Aliverti, A, Pandini, V, Fraaije, M.W, Heck, A.J.R, Edmondson, D.E, Mattevi, A. | | Deposit date: | 2012-10-01 | | Release date: | 2012-11-07 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Precursor of Ether Phospholipids is Synthesized by a Flavoenzyme Through Covalent Catalysis.
Proc.Natl.Acad.Sci.USA, 109, 2012
|
|
4BK2
 
 | | Crystal structure of 3-hydroxybenzoate 6-hydroxylase uncovers lipid- assisted flavoprotein strategy for regioselective aromatic hydroxylation: Q301E mutant | | Descriptor: | FLAVIN-ADENINE DINUCLEOTIDE, PHOSPHATIDYLGLYCEROL-PHOSPHOGLYCEROL, PROBABLE SALICYLATE MONOOXYGENASE | | Authors: | Orru, R, Montersino, S, Barendregt, A, Westphal, A.H, van Duijn, E, Mattevi, A, van Berkel, W.J.H. | | Deposit date: | 2013-04-21 | | Release date: | 2013-07-24 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.47 Å) | | Cite: | Crystal Structure of 3-Hydroxybenzoate 6-Hydroxylase Uncovers Lipid-Assisted Flavoprotein Strategy for Regioselective Aromatic Hydroxylation
J.Biol.Chem., 288, 2013
|
|
1H86
 
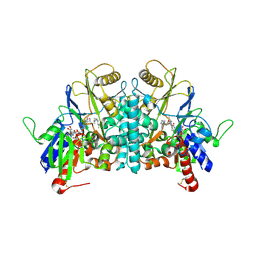 | | COVALENT ADDUCT BETWEEN POLYAMINE OXIDASE AND N1ethylN11((cycloheptyl)methyl)4,8diazaundecane at pH 7.0 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 3-[(3-{[3-(METHYLAMINO)PROPYL]AMINO}PROPYL)AMINO]PROPANE-1,1-DIOL, ... | | Authors: | Binda, C, Coda, A, Angelini, R, Federico, R, Ascenzi, P, Mattevi, A. | | Deposit date: | 2001-01-24 | | Release date: | 2001-01-31 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural Bases for Inhibitor Binding and Catalysis in Polyamine Oxidase
Biochemistry, 40, 2001
|
|
1H82
 
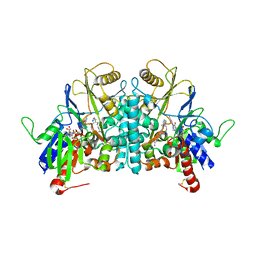 | | STRUCTURE OF POLYAMINE OXIDASE IN COMPLEX WITH GUAZATINE | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, FLAVIN-ADENINE DINUCLEOTIDE, N-{8-[(8-{[(E)-AMINO(IMINO)METHYL]AMINO}OCTYL)AMINO]OCTYL}GUANIDINE, ... | | Authors: | Binda, C, Coda, A, Angelini, R, Federico, R, Ascenzi, P, Mattevi, A. | | Deposit date: | 2001-01-24 | | Release date: | 2001-01-31 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structural Bases for Inhibitor Binding and Catalysis in Polyamine Oxidase
Biochemistry, 40, 2001
|
|
1H83
 
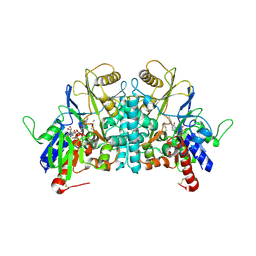 | | STRUCTURE OF POLYAMINE OXIDASE IN COMPLEX WITH 1,8-DIAMINOOCTANE | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, FLAVIN-ADENINE DINUCLEOTIDE, OCTANE 1,8-DIAMINE, ... | | Authors: | Binda, C, Coda, A, Angelini, R, Federico, R, Ascenzi, P, Mattevi, A. | | Deposit date: | 2001-01-24 | | Release date: | 2001-01-31 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structural Bases for Inhibitor Binding and Catalysis in Polyamine Oxidase
Biochemistry, 40, 2001
|
|
1H84
 
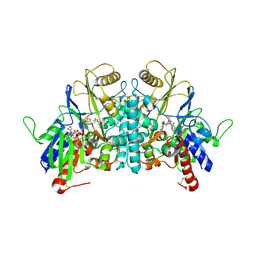 | | COVALENT ADDUCT BETWEEN POLYAMINE OXIDASE AND N1ethylN11((cycloheptyl)methyl)4,8diazaundecane at pH 4.6 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 3-[(3-{[3-(METHYLAMINO)PROPYL]AMINO}PROPYL)AMINO]PROPANE-1,1-DIOL, FLAVIN-ADENINE DINUCLEOTIDE, ... | | Authors: | Binda, C, Coda, A, Angelini, R, Federico, R, Ascenzi, P, Mattevi, A. | | Deposit date: | 2001-01-24 | | Release date: | 2001-01-31 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural Bases for Inhibitor Binding and Catalysis in Polyamine Oxidase
Biochemistry, 40, 2001
|
|
1H81
 
 | | STRUCTURE OF POLYAMINE OXIDASE IN THE REDUCED STATE | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, FLAVIN-ADENINE DINUCLEOTIDE, POLYAMINE OXIDASE, ... | | Authors: | Binda, C, Coda, A, Angelini, R, Federico, R, Ascenzi, P, Mattevi, A. | | Deposit date: | 2001-01-24 | | Release date: | 2001-01-31 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structural Bases for Inhibitor Binding and Catalysis in Polyamine Oxidase
Biochemistry, 40, 2001
|
|
