4R7X
 
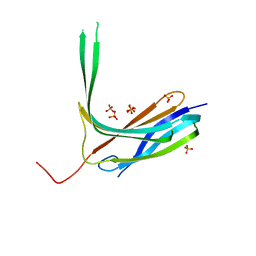 | | Crystal structure of N-lobe of human ARRDC3(1-180) | | Descriptor: | Arrestin domain-containing protein 3, PHOSPHATE ION | | Authors: | Qi, S, Hurley, J. | | Deposit date: | 2014-08-28 | | Release date: | 2014-10-01 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.61 Å) | | Cite: | Insights into beta 2-adrenergic receptor binding from structures of the N-terminal lobe of ARRDC3.
Protein Sci., 23, 2014
|
|
4R7V
 
 | | Crystal structure of N-lobe of human ARRDC3(1-165) | | Descriptor: | Arrestin domain-containing protein 3 | | Authors: | Qi, S, Hurley, J. | | Deposit date: | 2014-08-28 | | Release date: | 2014-10-01 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.73 Å) | | Cite: | Insights into beta 2-adrenergic receptor binding from structures of the N-terminal lobe of ARRDC3.
Protein Sci., 23, 2014
|
|
4GFX
 
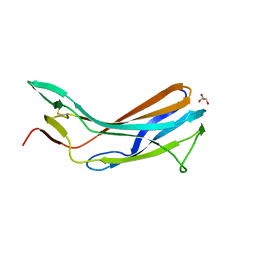 | | Crystal structure of the N-terminal domain of TXNIP | | Descriptor: | GLYCEROL, Thioredoxin-interacting protein | | Authors: | Hwang, J, Kim, M.H. | | Deposit date: | 2012-08-04 | | Release date: | 2014-02-05 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | The structural basis for the negative regulation of thioredoxin by thioredoxin-interacting protein.
Nat Commun, 5, 2014
|
|
4GEJ
 
 | | N-terminal domain of VDUP-1 | | Descriptor: | CALCIUM ION, Thioredoxin-interacting protein | | Authors: | Polekhina, G, Kok, S.F, Ascher, D.B, Waltham, M. | | Deposit date: | 2012-08-02 | | Release date: | 2013-02-27 | | Last modified: | 2017-11-15 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Structure of the N-terminal domain of human thioredoxin-interacting protein.
Acta Crystallogr.,Sect.D, 69, 2013
|
|
4GEI
 
 | | N-terminal domain of VDUP-1 | | Descriptor: | Thioredoxin-interacting protein | | Authors: | Polekhina, G, Kok, S.F, Ascher, D.B, Waltham, M. | | Deposit date: | 2012-08-02 | | Release date: | 2013-02-27 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Structure of the N-terminal domain of human thioredoxin-interacting protein.
Acta Crystallogr.,Sect.D, 69, 2013
|
|
1SUJ
 
 | |
1ZSH
 
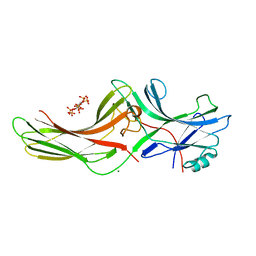 | | Crystal structure of bovine arrestin-2 in complex with inositol hexakisphosphate (IP6) | | Descriptor: | Beta-arrestin 1, INOSITOL HEXAKISPHOSPHATE, MAGNESIUM ION | | Authors: | Milano, S.K, Kim, Y.M, Stefano, F.P, Benovic, J.L, Brenner, C. | | Deposit date: | 2005-05-24 | | Release date: | 2006-01-31 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Nonvisual arrestin oligomerization and cellular localization are regulated by inositol hexakisphosphate binding
J.Biol.Chem., 281, 2006
|
|
1AYR
 
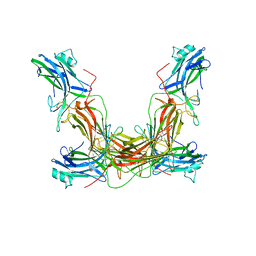 | | ARRESTIN FROM BOVINE ROD OUTER SEGMENTS | | Descriptor: | ARRESTIN | | Authors: | Granzin, J, Wilden, U, Choe, H.-W, Labahn, J, Krafft, B, Bueldt, G. | | Deposit date: | 1997-11-10 | | Release date: | 1998-11-25 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (3.3 Å) | | Cite: | X-ray crystal structure of arrestin from bovine rod outer segments.
Nature, 391, 1998
|
|
2WTR
 
 | |
1JSY
 
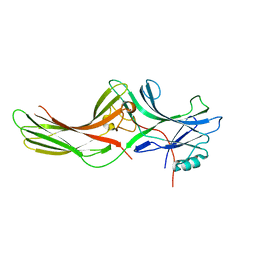 | | Crystal structure of bovine arrestin-2 | | Descriptor: | Bovine arrestin-2 (full length) | | Authors: | Milano, S.K, Pace, H.C, Kim, Y.M, Brenner, C, Benovic, J.L. | | Deposit date: | 2001-08-19 | | Release date: | 2002-03-27 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Scaffolding functions of arrestin-2 revealed by crystal structure and mutagenesis.
Biochemistry, 41, 2002
|
|
6BK9
 
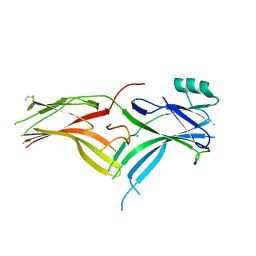 | | Crystal Structure of Squid Arrestin | | Descriptor: | CHLORIDE ION, Visual arrestin | | Authors: | Eger, B.T, Bandyopadhyay, A, Yedidi, R.S, Ernst, O.P. | | Deposit date: | 2017-11-08 | | Release date: | 2018-09-19 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (3.00005579 Å) | | Cite: | A Novel Polar Core and Weakly Fixed C-Tail in Squid Arrestin Provide New Insight into Interaction with Rhodopsin.
J. Mol. Biol., 430, 2018
|
|
6NI2
 
 | | Stabilized beta-arrestin 1-V2T subcomplex of a GPCR-G protein-beta-arrestin mega-complex | | Descriptor: | Beta-arrestin-1, Fab30 Heavy Chain, Fab30 Light Chain, ... | | Authors: | Nguyen, A.H, Thomsen, A.R.B, Cahill, T.J, des Georges, A, Lefkowitz, R.J. | | Deposit date: | 2018-12-26 | | Release date: | 2019-11-20 | | Last modified: | 2019-12-18 | | Method: | ELECTRON MICROSCOPY (4 Å) | | Cite: | Structure of an endosomal signaling GPCR-G protein-beta-arrestin megacomplex.
Nat.Struct.Mol.Biol., 26, 2019
|
|
3GC3
 
 | | Crystal Structure of Arrestin2S and Clathrin | | Descriptor: | Beta-arrestin-1, Clathrin heavy chain 1 | | Authors: | Williams, J.C, Kang, D.S. | | Deposit date: | 2009-02-21 | | Release date: | 2009-08-25 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structure of an arrestin2-clathrin complex reveals a novel clathrin binding domain that modulates receptor trafficking.
J.Biol.Chem., 284, 2009
|
|
3GD1
 
 | | Structure of an Arrestin/Clathrin complex reveals a novel clathrin binding domain that modulates receptor trafficking | | Descriptor: | Beta-arrestin-1, Clathrin heavy chain 1, clathrin | | Authors: | Kang, D.S, Kern, R.C, Puthenveedu, M.A, von Zastrow, M, Williams, J.C, Benovic, J.L. | | Deposit date: | 2009-02-23 | | Release date: | 2009-08-25 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (3.5 Å) | | Cite: | Structure of an arrestin2-clathrin complex reveals a novel clathrin binding domain that modulates receptor trafficking
J.Biol.Chem., 284, 2009
|
|
7R0J
 
 | | Structure of the V2 receptor Cter-arrestin2-ScFv30 complex | | Descriptor: | Arrestin2, ScFv30, V2R Cter | | Authors: | Bous, J, Fouillen, A, Trapani, S, Granier, S, Mouillac, B, Bron, P. | | Deposit date: | 2022-02-02 | | Release date: | 2022-09-14 | | Method: | ELECTRON MICROSCOPY (4.23 Å) | | Cite: | Structure of the vasopressin hormone-V2 receptor-beta-arrestin1 ternary complex.
Sci Adv, 8, 2022
|
|
5TV1
 
 | | active arrestin-3 with inositol hexakisphosphate | | Descriptor: | Beta-arrestin-2, GLYCEROL, INOSITOL HEXAKISPHOSPHATE | | Authors: | Chen, Q, Gilbert, N.C, Perry, N.A, Vishniveteskiy, S, Gurevich, V.V, Iverson, T.M. | | Deposit date: | 2016-11-07 | | Release date: | 2017-11-22 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structural basis of arrestin-3 activation and signaling.
Nat Commun, 8, 2017
|
|
6K3F
 
 | |
7USY
 
 | | Structure of C. elegans TMC-1 complex with ARRD-6 | | Descriptor: | 1,2-Distearoyl-sn-glycerophosphoethanolamine, 1,2-dioleoyl-sn-glycero-3-phosphoethanolamine, 2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Jeong, H, Clark, S, Gouaux, E. | | Deposit date: | 2022-04-26 | | Release date: | 2022-10-19 | | Last modified: | 2023-08-09 | | Method: | ELECTRON MICROSCOPY (3.54 Å) | | Cite: | Structures of the TMC-1 complex illuminate mechanosensory transduction.
Nature, 610, 2022
|
|
4ZRG
 
 | | Visual arrestin mutant - R175E | | Descriptor: | CARBON DIOXIDE, S-arrestin | | Authors: | Granzin, J, Stadler, A, Cousin, A, Schlesinger, R, Batra-Safferling, R. | | Deposit date: | 2015-05-12 | | Release date: | 2015-11-11 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Structural evidence for the role of polar core residue Arg175 in arrestin activation.
Sci Rep, 5, 2015
|
|
3UGU
 
 | |
3UGX
 
 | | Crystal Structure of Visual Arrestin | | Descriptor: | 1,2-ETHANEDIOL, IMIDAZOLE, PENTANEDIAL, ... | | Authors: | Batra-Safferling, R, Granzin, J. | | Deposit date: | 2011-11-03 | | Release date: | 2012-02-08 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.649 Å) | | Cite: | Crystal Structure of p44, a Constitutively Active Splice Variant of Visual Arrestin.
J.Mol.Biol., 416, 2012
|
|
6KL7
 
 | | Beta-arrestin 1 mutant S13D/T275D | | Descriptor: | 1,2-ETHANEDIOL, BARIUM ION, Beta-arrestin-1 | | Authors: | Kang, H, Choi, H.J. | | Deposit date: | 2019-07-29 | | Release date: | 2020-01-29 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.794 Å) | | Cite: | Conformational Dynamics and Functional Implications of Phosphorylated beta-Arrestins.
Structure, 28, 2020
|
|
8AS3
 
 | | Structure of arrestin2 in complex with 6P CCR5 phosphopeptide and Fab30 | | Descriptor: | Beta-arrestin-1, C-C chemokine receptor type 5, Fab30 heavy chain, ... | | Authors: | Isaikina, P, Jakob, R.P, Maier, T, Grzesiek, S. | | Deposit date: | 2022-08-18 | | Release date: | 2023-06-07 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (3.5 Å) | | Cite: | A key GPCR phosphorylation motif discovered in arrestin2⋅CCR5 phosphopeptide complexes.
Mol.Cell, 83, 2023
|
|
8AS4
 
 | |
8AS2
 
 | | Structure of arrestin2 in complex with 4P CCR5 phosphopeptide and Fab30 | | Descriptor: | Beta-arrestin-1, C-C chemokine receptor type 5, Fab30 heavy chain, ... | | Authors: | Isaikina, P, Jakob, R.P, Maier, T, Grzesiek, S. | | Deposit date: | 2022-08-18 | | Release date: | 2023-06-07 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | A key GPCR phosphorylation motif discovered in arrestin2⋅CCR5 phosphopeptide complexes.
Mol.Cell, 83, 2023
|
|
