9QLM
 
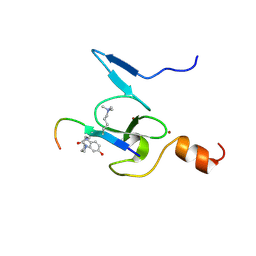 | | Solution structure of the TAF3-PHD bound to a H3K4me3Q5ser histone tail peptide with a serotonylated glutamine | | Descriptor: | Histone H3.1, SEROTONIN, Transcription initiation factor TFIID subunit 3, ... | | Authors: | van Ingen, H, Gielingh, H, Pulido-Cortes, L, Thijssen, V, Timmers, H.T.M, Jongkees, S, Honorato, R.V, Bonvin, A.M.J.J, Liu, M, Yoshisada, R, Soares, L.R. | | Deposit date: | 2025-03-21 | | Release date: | 2025-05-14 | | Last modified: | 2025-07-23 | | Method: | SOLUTION NMR | | Cite: | Molecular determinants for recognition of serotonylated chromatin.
Nucleic Acids Res., 53, 2025
|
|
8VLF
 
 | |
8VLD
 
 | |
8VLH
 
 | |
9EL8
 
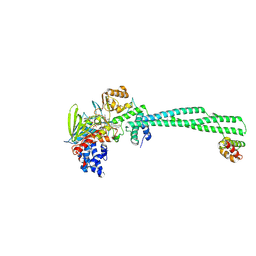 | | LSD1-CoREST in complex with T105 1S2R enantiomer | | Descriptor: | 3-[(1R,2S)-2-(cyclobutylamino)cyclopropyl]-N-(5-methyl-1,3,4-thiadiazol-2-yl)benzamide, Lysine-specific histone demethylase 1A, REST corepressor 1, ... | | Authors: | Caroli, J, Mattevi, A. | | Deposit date: | 2024-12-04 | | Release date: | 2025-02-05 | | Method: | X-RAY DIFFRACTION (2.91 Å) | | Cite: | Distal drug resistance mutations promote covalent inhibitor-adduct Grob fragmentation in LSD1
to be published
|
|
9ELA
 
 | | LSD1-CoREST in complex with T108, long soaking | | Descriptor: | Lysine-specific histone demethylase 1A, REST corepressor 1, [(2R,3S,4R,5R)-5-(6-amino-9H-purin-9-yl)-3,4-dihydroxyoxolan-2-yl]methyl (2R,3S,4S)-2,3,4-trihydroxy-5-[(1R,3R,3aS,13R)-1-hydroxy-10,11-dimethyl-4,6-dioxo-3-[3-(phenylcarbamoyl)phenyl]-2,3,5,6-tetrahydro-1H-benzo[g]pyrrolo[2,1-e]pteridin-8(4H)-yl]pentyl dihydrogen diphosphate | | Authors: | Caroli, J, Mattevi, A. | | Deposit date: | 2024-12-04 | | Release date: | 2025-02-05 | | Method: | X-RAY DIFFRACTION (2.85 Å) | | Cite: | Distal drug resistance mutations promote covalent inhibitor-adduct Grob fragmentation in LSD1
to be published
|
|
9EL7
 
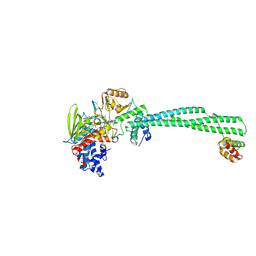 | | LSD1-CoREST in complex with T105 enantiomer (1R,2S) | | Descriptor: | Lysine-specific histone demethylase 1A, REST corepressor 1, [(2R,3S,4R,5R)-5-(6-amino-9H-purin-9-yl)-3,4-dihydroxyoxolan-2-yl]methyl (2S,3S,4R)-5-[(4aS)-7,8-dimethyl-4a-[(1R)-1-{3-[(5-methyl-1,3,4-thiadiazol-2-yl)carbamoyl]phenyl}-3-oxopropyl]-2,4-dioxo-3,4,4a,5-tetrahydrobenzo[g]pteridin-10(2H)-yl]-2,3,4-trihydroxypentyl dihydrogen diphosphate (non-preferred name) | | Authors: | Caroli, J, Mattevi, A. | | Deposit date: | 2024-12-04 | | Release date: | 2025-02-05 | | Method: | X-RAY DIFFRACTION (2.87 Å) | | Cite: | Distal drug resistance mutations promote covalent inhibitor-adduct Grob fragmentation in LSD1
to be published
|
|
8ULC
 
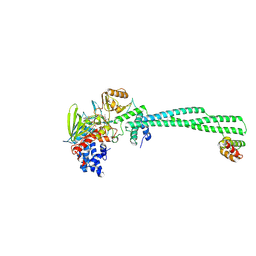 | | LSD1-CoREST in complex with T15, long soaking | | Descriptor: | Lysine-specific histone demethylase 1A, REST corepressor 1, methyl 3-{(1R,3S,3aS,13R)-8-[(2S,3S,4R)-5-{[(S)-{[(S)-{[(2R,3S,4R,5R)-5-(6-amino-9H-purin-9-yl)-3,4-dihydroxyoxolan-2-yl]methoxy}(hydroxy)phosphoryl]oxy}(hydroxy)phosphoryl]oxy}-2,3,4-trihydroxypentyl]-1-hydroxy-10,11-dimethyl-4,6-dioxo-2,3,4,5,6,8-hexahydro-1H-benzo[g]pyrrolo[2,1-e]pteridin-3-yl}benzoate (non-preferred name) | | Authors: | Caroli, J, Mattevi, A. | | Deposit date: | 2023-10-16 | | Release date: | 2025-02-05 | | Method: | X-RAY DIFFRACTION (2.82 Å) | | Cite: | Distal drug resistance mutations promote covalent inhibitor-adduct Grob fragmentation in LSD1
To Be Published
|
|
8UL8
 
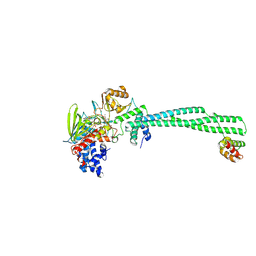 | | LSD1-CoREST in complex with T15, short soaking | | Descriptor: | Lysine-specific histone demethylase 1A, REST corepressor 1, methyl 3-{(1R,3S,3aS,13R)-8-[(2S,3S,4R)-5-{[(S)-{[(S)-{[(2R,3S,4R,5R)-5-(6-amino-9H-purin-9-yl)-3,4-dihydroxyoxolan-2-yl]methoxy}(hydroxy)phosphoryl]oxy}(hydroxy)phosphoryl]oxy}-2,3,4-trihydroxypentyl]-1-hydroxy-10,11-dimethyl-4,6-dioxo-2,3,4,5,6,8-hexahydro-1H-benzo[g]pyrrolo[2,1-e]pteridin-3-yl}benzoate (non-preferred name) | | Authors: | Caroli, J, Mattevi, A. | | Deposit date: | 2023-10-16 | | Release date: | 2025-02-05 | | Method: | X-RAY DIFFRACTION (2.82 Å) | | Cite: | Distal drug resistance mutations promote covalent inhibitor-adduct Grob fragmentation in LSD1
To Be Published
|
|
8UL6
 
 | | LSD1-CoREST in complex with T16, long soaking | | Descriptor: | Lysine-specific histone demethylase 1A, REST corepressor 1, [(2R,3S,4R,5R)-5-(6-amino-9H-purin-9-yl)-3,4-dihydroxyoxolan-2-yl]methyl (2R,3S,4S)-2,3,4-trihydroxy-5-[(1R,3S,3aS,13R)-1-hydroxy-10,11-dimethyl-3-[3-(methylcarbamoyl)phenyl]-4,6-dioxo-2,3,5,6-tetrahydro-1H-benzo[g]pyrrolo[2,1-e]pteridin-8(4H)-yl]pentyl dihydrogen diphosphate (non-preferred name) | | Authors: | Caroli, J, Mattevi, A. | | Deposit date: | 2023-10-16 | | Release date: | 2025-02-05 | | Method: | X-RAY DIFFRACTION (2.74 Å) | | Cite: | Distal drug resistance mutations promote covalent inhibitor-adduct Grob fragmentation in LSD1
To Be Published
|
|
8UNI
 
 | |
8UOM
 
 | |
8ULB
 
 | | LSD1-CoREST in complex with T17, long soaking | | Descriptor: | Lysine-specific histone demethylase 1A, REST corepressor 1, [(2R,3S,4R,5R)-5-(6-amino-9H-purin-9-yl)-3,4-dihydroxyoxolan-2-yl]methyl (2R,3S,4S)-5-[(1R,3S,3aS,13R)-3-[3-(dimethylcarbamoyl)phenyl]-1-hydroxy-10,11-dimethyl-4,6-dioxo-2,3,5,6-tetrahydro-1H-benzo[g]pyrrolo[2,1-e]pteridin-8(4H)-yl]-2,3,4-trihydroxypentyl dihydrogen diphosphate (non-preferred name) | | Authors: | Caroli, J, Mattevi, A. | | Deposit date: | 2023-10-16 | | Release date: | 2025-02-05 | | Method: | X-RAY DIFFRACTION (3.41 Å) | | Cite: | Distal drug resistance mutations promote covalent inhibitor-adduct Grob fragmentation in LSD1
To Be Published
|
|
8UMQ
 
 | | LSD1-CoREST in complex with T18, long soaking | | Descriptor: | Lysine-specific histone demethylase 1A, REST corepressor 1, [(2R,3S,4R,5R)-5-(6-amino-9H-purin-9-yl)-3,4-dihydroxyoxolan-2-yl]methyl (2R,3S,4S)-5-[(1R,3S,3aS,13R)-3-(3-benzamidophenyl)-1-hydroxy-10,11-dimethyl-4,6-dioxo-2,3,5,6-tetrahydro-1H-benzo[g]pyrrolo[2,1-e]pteridin-8(4H)-yl]-2,3,4-trihydroxypentyl dihydrogen diphosphate (non-preferred name) | | Authors: | Caroli, J, Mattevi, A. | | Deposit date: | 2023-10-18 | | Release date: | 2025-02-05 | | Method: | X-RAY DIFFRACTION (3.26 Å) | | Cite: | Distal drug resistance mutations promote covalent inhibitor-adduct Grob fragmentation in LSD1
To Be Published
|
|
8GJ6
 
 | | LSD1-CoREST in complex with T16 | | Descriptor: | Lysine-specific histone demethylase 1A, POTASSIUM ION, REST corepressor 1, ... | | Authors: | Caroli, J, Mattevi, A. | | Deposit date: | 2023-03-15 | | Release date: | 2025-02-05 | | Method: | X-RAY DIFFRACTION (2.77 Å) | | Cite: | Distal drug resistance mutations promote covalent inhibitor-adduct Grob fragmentation in LSD1
To Be Published
|
|
8VMN
 
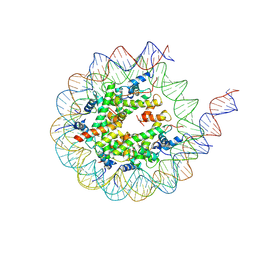 | | H3K4me3 nucleosome bound to PRC2_AJ1-450 | | Descriptor: | DNA (157-MER), Histone H2A, Histone H2B, ... | | Authors: | Cookis, T, Nogales, E. | | Deposit date: | 2024-01-13 | | Release date: | 2025-01-22 | | Last modified: | 2025-03-05 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | Structural basis for the inhibition of PRC2 by active transcription histone posttranslational modifications.
Nat.Struct.Mol.Biol., 32, 2025
|
|
8VO0
 
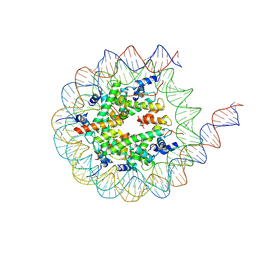 | |
8VNV
 
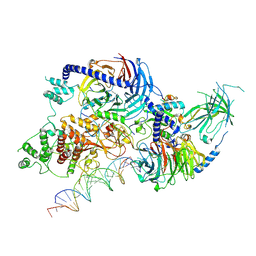 | |
8VMJ
 
 | | H3K4me3 nucleosome bound to PRC2_AJ119-450 | | Descriptor: | DNA (157-MER), Histone H2A, Histone H2B, ... | | Authors: | Cookis, T, Nogales, E. | | Deposit date: | 2024-01-13 | | Release date: | 2025-01-15 | | Last modified: | 2025-03-05 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Structural basis for the inhibition of PRC2 by active transcription histone posttranslational modifications.
Nat.Struct.Mol.Biol., 32, 2025
|
|
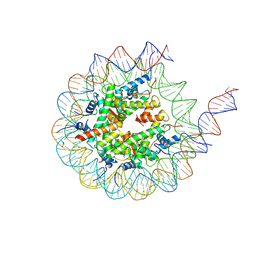
8VML
 
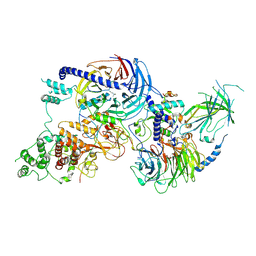 | | PRC2_AJ1-450 bound to H3K4me3 | | Descriptor: | AEPB2, EED, EZH2, ... | | Authors: | Cookis, T, Nogales, E. | | Deposit date: | 2024-01-13 | | Release date: | 2025-01-15 | | Last modified: | 2025-03-05 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | Structural basis for the inhibition of PRC2 by active transcription histone posttranslational modifications.
Nat.Struct.Mol.Biol., 32, 2025
|
|
8VOB
 
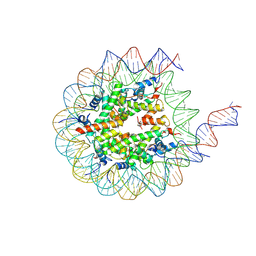 | | H3K36me3-modified nucleosome bound to PRC2_AJ1-450 | | Descriptor: | DNA (157-MER), Histone H2A type 1, Histone H2B 1.1, ... | | Authors: | Cookis, T, Nogales, E. | | Deposit date: | 2024-01-14 | | Release date: | 2025-01-15 | | Last modified: | 2025-03-05 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Structural basis for the inhibition of PRC2 by active transcription histone posttranslational modifications.
Nat.Struct.Mol.Biol., 32, 2025
|
|
8VMI
 
 | | PRC2_AJ119-450 bound to H3K4me3 | | Descriptor: | EZH2, Histone H3.1, Histone H3.1t, ... | | Authors: | Cookis, T, Nogales, E. | | Deposit date: | 2024-01-13 | | Release date: | 2025-01-15 | | Last modified: | 2025-05-28 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Structural basis for the inhibition of PRC2 by active transcription histone posttranslational modifications.
Nat.Struct.Mol.Biol., 32, 2025
|
|
8VNZ
 
 | |
9GP4
 
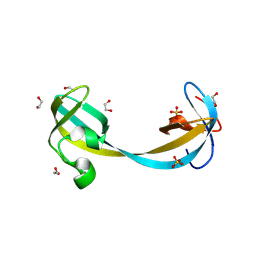 | | Jumonji domain-containing protein 2A with crystallization epitope mutations Q953E:A958D | | Descriptor: | 1,2-ETHANEDIOL, Lysine-specific demethylase 4A, SULFATE ION | | Authors: | Fairhead, M, Strain-Damerell, C, Ye, M, Mackinnon, S.R, Pinkas, D, MacLean, E.M, Koekemoer, L, Damerell, D, Krojer, T, Arrowsmith, C.H, Edwards, A, Bountra, C, Yue, W, Burgess-Brown, N, Marsden, B, von Delft, F, Structural Genomics Consortium (SGC) | | Deposit date: | 2024-09-06 | | Release date: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.59 Å) | | Cite: | A fast, parallel method for efficiently exploring crystallization behaviour of large numbers of protein variants
To be published
|
|
9GD2
 
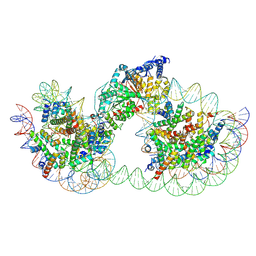 | | Structure of Chd1 bound to a dinucleosome with a dyad-to-dyad distance of 103 bp. | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, BERYLLIUM TRIFLUORIDE ION, Chromo domain-containing protein 1, ... | | Authors: | Engeholm, M, Roske, J.J, Oberbeckmann, E, Dienemann, C, Lidschreiber, M, Cramer, P, Farnung, L. | | Deposit date: | 2024-08-04 | | Release date: | 2024-09-18 | | Last modified: | 2024-10-02 | | Method: | ELECTRON MICROSCOPY (4.2 Å) | | Cite: | Resolution of transcription-induced hexasome-nucleosome complexes by Chd1 and FACT.
Mol.Cell, 84, 2024
|
|
