5OGR
 
 | | Structure of cathepsin B1 from Schistosoma mansoni in complex with WRR286 inhibitor | | Descriptor: | 3-[[N-[4-METHYL-PIPERAZINYL]CARBONYL]-PHENYLALANINYL-AMINO]-5-PHENYL-PENTANE-1-SULFONIC ACID BENZYLOXY-AMIDE, ACETATE ION, Cathepsin B-like peptidase (C01 family) | | Authors: | Jilkova, A, Rezacova, P, Brynda, J, Mares, M. | | Deposit date: | 2017-07-13 | | Release date: | 2018-11-21 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Druggable Hot Spots in the Schistosomiasis Cathepsin B1 Target Identified by Functional and Binding Mode Analysis of Potent Vinyl Sulfone Inhibitors.
Acs Infect Dis., 2020
|
|
5OGQ
 
 | | Structure of cathepsin B1 from Schistosoma mansoni in complex with WRR391 inhibitor | | Descriptor: | ACETATE ION, Cathepsin B-like peptidase (C01 family), ethyl 1-[[(2~{S})-3-(4-hydroxyphenyl)-1-oxidanylidene-1-[[(3~{S})-1-phenyl-5-pyridin-2-ylsulfonyl-pentan-3-yl]amino]propan-2-yl]carbamoyl]piperidine-4-carboxylate | | Authors: | Jilkova, A, Rezacova, P, Brynda, J, Mares, M. | | Deposit date: | 2017-07-13 | | Release date: | 2018-11-21 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.91 Å) | | Cite: | Druggable Hot Spots in the Schistosomiasis Cathepsin B1 Target Identified by Functional and Binding Mode Analysis of Potent Vinyl Sulfone Inhibitors.
Acs Infect Dis., 2020
|
|
5MQY
 
 | | CATHEPSIN L IN COMPLEX WITH 4-[1,3-benzodioxol-5-ylmethyl(2-phenoxyethyl)amino]-5-fluoropyrimidine-2-carbonitrile | | Descriptor: | 1,2-ETHANEDIOL, 4-[1,3-benzodioxol-5-ylmethyl(2-phenoxyethyl)amino]-5-fluoropyrimidine-2-carbonitrile, Cathepsin L1 | | Authors: | Kuglstatter, A, Stihle, M, Benz, J. | | Deposit date: | 2016-12-21 | | Release date: | 2017-03-22 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.13 Å) | | Cite: | Prospective Evaluation of Free Energy Calculations for the Prioritization of Cathepsin L Inhibitors.
J. Med. Chem., 60, 2017
|
|
5MBM
 
 | |
5MBL
 
 | |
5MAJ
 
 | | CATHEPSIN L IN COMPLEX WITH 4-[cyclopentyl(imidazo[1,2-a]pyridin-2-ylmethyl)amino]-6-morpholino-1,3,5-triazine-2-carbonitrile | | Descriptor: | 1,2-ETHANEDIOL, Cathepsin L1, ~{N}-cyclopentyl-~{N}-(imidazo[1,2-a]pyridin-2-ylmethyl)-4-(iminomethyl)-6-morpholin-4-yl-1,3,5-triazin-2-amine | | Authors: | Kuglstatter, A, Stihle, M, Benz, J. | | Deposit date: | 2016-11-03 | | Release date: | 2017-01-11 | | Last modified: | 2017-02-22 | | Method: | X-RAY DIFFRACTION (1 Å) | | Cite: | Inhibition of the Cysteine Protease Human Cathepsin L by Triazine Nitriles: AmideHeteroarene pi-Stacking Interactions and Chalcogen Bonding in the S3 Pocket.
ChemMedChem, 12, 2017
|
|
5MAE
 
 | | CATHEPSIN L IN COMPLEX WITH (2S,4R)-4-(2-Chloro-4-methoxy-benzenesulfonyl)-1-[3-(5-chloro-pyridin-2-yl)-azetidine-3-carbonyl]-pyrrolidine-2-car boxylic acid (1-cyano-cyclopropyl)-amide | | Descriptor: | 1,2-ETHANEDIOL, Cathepsin L1, [4-[cyclopentyl(pyrazin-2-ylmethyl)amino]-6-morpholin-4-yl-1,3,5-triazin-2-yl]methylideneazanide | | Authors: | Kuglstatter, A, Stihle, M, Benz, J. | | Deposit date: | 2016-11-03 | | Release date: | 2017-01-11 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (1 Å) | | Cite: | Inhibition of the Cysteine Protease Human Cathepsin L by Triazine Nitriles: AmideHeteroarene pi-Stacking Interactions and Chalcogen Bonding in the S3 Pocket.
ChemMedChem, 12, 2017
|
|
5JT8
 
 | | Structural basis for the limited antibody cross reactivity between the mite allergens Blo t 1 and Der p 1 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Blo t 1 allergen, ... | | Authors: | Meno, K, Kastrup, J.S, Gajhede, M. | | Deposit date: | 2016-05-09 | | Release date: | 2017-05-24 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | The structure of the mite allergen Blo t 1 explains the limited antibody cross-reactivity to Der p 1.
Allergy, 72, 2017
|
|
5JH3
 
 | | Human cathepsin K mutant C25S | | Descriptor: | ACETATE ION, CHLORIDE ION, Cathepsin K, ... | | Authors: | Novinec, M, Korenc, M, Lenarcic, B. | | Deposit date: | 2016-04-20 | | Release date: | 2016-11-30 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | An allosteric site enables fine-tuning of cathepsin K by diverse effectors.
FEBS Lett., 590, 2016
|
|
5JA7
 
 | | Human cathepsin K mutant C25S in complex with the allosteric effector NSC94914 | | Descriptor: | ACETATE ION, Cathepsin K, GLYCEROL, ... | | Authors: | Novinec, M, Korenc, M, Lenarcic, B. | | Deposit date: | 2016-04-12 | | Release date: | 2016-11-30 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.61 Å) | | Cite: | An allosteric site enables fine-tuning of cathepsin K by diverse effectors.
FEBS Lett., 590, 2016
|
|
5J94
 
 | | Human cathepsin K mutant C25S in complex with the allosteric effector NSC13345 | | Descriptor: | 2-{[(carbamoylsulfanyl)acetyl]amino}benzoic acid, Cathepsin K, SULFATE ION | | Authors: | Novinec, M, Korenc, M, Lenarcic, B, Baici, A. | | Deposit date: | 2016-04-08 | | Release date: | 2016-04-20 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.22002459 Å) | | Cite: | A novel allosteric mechanism in the cysteine peptidase cathepsin K discovered by computational methods.
Nat Commun, 5, 2014
|
|
5I4H
 
 | |
5FPW
 
 | |
5F02
 
 | | CATHEPSIN L IN COMPLEX WITH (2S,4R)-4-(2-Chloro-4-methoxy-benzenesulfonyl)-1-[3-(5-chloro-pyridin-2-yl)-azetidine-3-carbonyl]-pyrrolidine-2-carboxylic acid (1-cyano-cyclopropyl)-amide | | Descriptor: | (2~{S},4~{R})-4-[(2-chloranyl-4-methoxy-phenyl)-bis(oxidanyl)-$l^{4}-sulfanyl]-1-[3-(5-chloranylpyridin-2-yl)azetidin-3-yl]carbonyl-~{N}-[1-(iminomethyl)cyclopropyl]pyrrolidine-2-carboxamide, Cathepsin L1, GLYCEROL | | Authors: | Banner, D, Benz, J, Stihle, M, Kuglstatter, A. | | Deposit date: | 2015-11-27 | | Release date: | 2016-02-24 | | Last modified: | 2016-05-25 | | Method: | X-RAY DIFFRACTION (1.43 Å) | | Cite: | A Real-World Perspective on Molecular Design.
J.Med.Chem., 59, 2016
|
|
5EGW
 
 | |
5EF4
 
 | | 2.05 A crystal structure of the Amb a 11 cysteine protease, a major ragweed pollen allergen, in its proform | | Descriptor: | Cysteine protease, GLYCEROL | | Authors: | Briozzo, P, Kopecny, D, Savko, M. | | Deposit date: | 2015-10-23 | | Release date: | 2016-04-27 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Structural and Functional Characterization of the Major Allergen Amb a 11 from Short Ragweed Pollen.
J.Biol.Chem., 291, 2016
|
|
5A24
 
 | | Crystal structure of Dionain-1, the major endopeptidase in the Venus flytrap digestive juice | | Descriptor: | DIONAIN-1, N-[N-[1-HYDROXYCARBOXYETHYL-CARBONYL]LEUCYLAMINO-BUTYL]-GUANIDINE, PHOSPHATE ION | | Authors: | Risor, M.W, Thomsen, L.R, Sanggaard, K.W, Nielsen, T.A, Thogersen, I.B, Lukassen, M.V, Rossen, L, Garcia-Ferrer, I, Guevara, T, Meinjohanns, E, Nielsen, N.C, Gomis-Ruth, F.X, Enghild, J.J. | | Deposit date: | 2015-05-12 | | Release date: | 2015-12-09 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Enzymatic and Structural Characterization of the Major Endopeptidase in the Venus Flytrap Digestion Fluid.
J.Biol.Chem., 291, 2016
|
|
4YYW
 
 | | Ficin D2 | | Descriptor: | Ficin isoform D, SULFATE ION | | Authors: | Azarkan, M, Baeyens-Volant, D, Loris, R. | | Deposit date: | 2015-03-24 | | Release date: | 2016-04-13 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.446 Å) | | Cite: | Crystal structure of four variants of the protease Ficin from the common fig
To Be Published
|
|
4YYV
 
 | | Ficin isoform C crystal form II | | Descriptor: | 3,6,9,12,15,18-HEXAOXAICOSANE, Ficin isoform C, SULFATE ION | | Authors: | Azarkan, M, Baeyens-Volant, D, Loris, R. | | Deposit date: | 2015-03-24 | | Release date: | 2016-04-13 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.902 Å) | | Cite: | Crystal structure of four variants of the protease Ficin from the common fig
To Be Published
|
|
4YYU
 
 | | Ficin C crystal form I | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, Ficin isoform C, SULFATE ION | | Authors: | Azarkan, M, Baeyens-Volant, D, Loris, R. | | Deposit date: | 2015-03-24 | | Release date: | 2016-04-13 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.177 Å) | | Cite: | Crystal structure of four variants of the protease Ficin from the common fig
To Be Published
|
|
4YYS
 
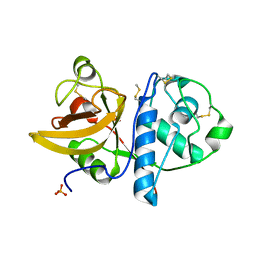 | | Ficin B crystal form II | | Descriptor: | Ficin isoform B, PHOSPHATE ION | | Authors: | Azarkan, M, Baeyens-Volant, D, Loris, R. | | Deposit date: | 2015-03-24 | | Release date: | 2016-04-13 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.35 Å) | | Cite: | Crystal structure of four variants of the protease Ficin from the common fig
To Be Published
|
|
4YYR
 
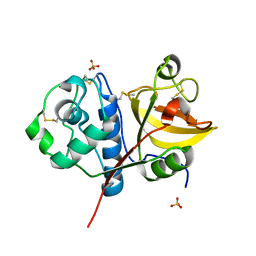 | | Ficin B crystal form I | | Descriptor: | Ficin B, SULFATE ION | | Authors: | Azarkan, M, Baeyens-Volant, D, Loris, R. | | Deposit date: | 2015-03-24 | | Release date: | 2016-04-13 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.349 Å) | | Cite: | Crystal structure of four variants of the protease Ficin from the common fig
To Be Published
|
|
4YYQ
 
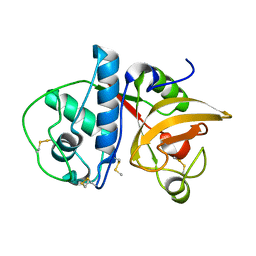 | | Ficin A | | Descriptor: | Ficin isoform A | | Authors: | Azarkan, M, Baeyens-Volant, D, Loris, R. | | Deposit date: | 2015-03-24 | | Release date: | 2016-04-13 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.594 Å) | | Cite: | Crystal structure of four variants of the protease Ficin from the common fig
To Be Published
|
|
4YVA
 
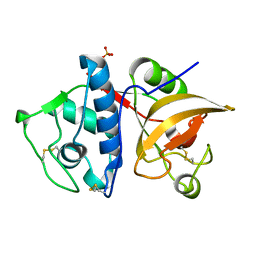 | | Cathepsin K co-crystallized with actinomycetes extract | | Descriptor: | Cathepsin K, SULFATE ION | | Authors: | Aguda, A.H, Nguyen, N.T, Bromme, D, Brayer, G.D. | | Deposit date: | 2015-03-19 | | Release date: | 2016-05-04 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Affinity Crystallography: A New Approach to Extracting High-Affinity Enzyme Inhibitors from Natural Extracts.
J.Nat.Prod., 79, 2016
|
|
4YV8
 
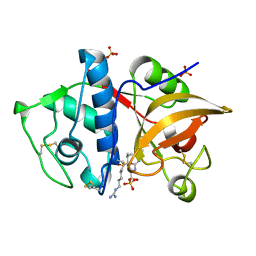 | | Crystal structure of cathepsin K bound to the covalent inhibitor lichostatinal | | Descriptor: | Cathepsin K, Lichostatinal, SULFATE ION | | Authors: | Aguda, A.H, Nguyen, N.T, Bromme, D, Brayer, G.D. | | Deposit date: | 2015-03-19 | | Release date: | 2016-05-04 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Affinity Crystallography: A New Approach to Extracting High-Affinity Enzyme Inhibitors from Natural Extracts.
J.Nat.Prod., 79, 2016
|
|
