7B73
 
 | | Insight into the molecular determinants of thermal stability in halohydrin dehalogenase HheD2. | | Descriptor: | 2-[N-CYCLOHEXYLAMINO]ETHANE SULFONIC ACID, GLYCEROL, Short-chain dehydrogenase/reductase SDR | | Authors: | Wessel, J, Petrillo, G, Estevez, M, Bosh, S, Seeger, M, Dijkman, W.P, Hidalgo, A, Uson, I, Osuna, S, Schallmey, A. | | Deposit date: | 2020-12-09 | | Release date: | 2021-04-07 | | Last modified: | 2024-06-19 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Insights into the molecular determinants of thermal stability in halohydrin dehalogenase HheD2.
Febs J., 288, 2021
|
|
2AKQ
 
 | | The structure of bovine B-lactoglobulin A in crystals grown at very low ionic strength | | Descriptor: | Beta-lactoglobulin variant A | | Authors: | Adams, J.J, Anderson, B.F, Norris, G.E, Creamer, L.K, Jameson, G.B. | | Deposit date: | 2005-08-03 | | Release date: | 2005-08-16 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Structure of bovine beta-lactoglobulin (variant A) at very low ionic strength
J.Struct.Biol., 154, 2006
|
|
5XGT
 
 | |
6EX7
 
 | | Crystal structure of NDM-1 metallo-beta-lactamase in complex with Cd ions and a hydrolyzed beta-lactam ligand - new refinement | | Descriptor: | 1,2-ETHANEDIOL, 2-[2-(2-METHOXY-ETHOXY)-ETHOXY]-ETHOXYL, CADMIUM ION, ... | | Authors: | Kim, Y, Raczynska, J.E, Shabalin, I.G, Jaskolski, M, Minor, W, Wlodawer, A, Tesar, C, Jedrzejczak, R, Babnigg, J, Mire, J, Sacchettini, J, Joachimiak, A. | | Deposit date: | 2017-11-07 | | Release date: | 2017-12-13 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | A close look onto structural models and primary ligands of metallo-beta-lactamases.
Drug Resist. Updat., 40, 2018
|
|
5XX2
 
 | | A BPTI-[5,55] variant with C14GA38L mutations | | Descriptor: | Pancreatic trypsin inhibitor, SULFATE ION | | Authors: | Islam, M.M. | | Deposit date: | 2017-07-01 | | Release date: | 2018-07-04 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.12 Å) | | Cite: | Hydrophobic surface residues can stabilize a protein through improved water-protein interactions.
Febs J., 2019
|
|
5XX4
 
 | | A BPTI-[5,55] variant with C14GA38K mutations | | Descriptor: | Pancreatic trypsin inhibitor, SULFATE ION | | Authors: | Islam, M.M. | | Deposit date: | 2017-07-01 | | Release date: | 2018-07-04 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.67 Å) | | Cite: | Hydrophobic surface residues can stabilize a protein through improved water-protein interactions.
Febs J., 2019
|
|
5XX5
 
 | | A BPTI-[5,55] variant with C14GA38I mutations | | Descriptor: | Pancreatic trypsin inhibitor, SULFATE ION | | Authors: | Islam, M.M. | | Deposit date: | 2017-07-01 | | Release date: | 2018-07-04 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.38 Å) | | Cite: | Hydrophobic surface residues can stabilize a protein through improved water-protein interactions.
Febs J., 2019
|
|
2KQO
 
 | |
6P90
 
 | | Crystal structure of PaDHDPS2-H56Q mutant | | Descriptor: | 4-hydroxy-tetrahydrodipicolinate synthase, CHLORIDE ION, GLYCEROL | | Authors: | Impey, R.E, Panjikar, S, Hall, C.J, Bock, L.J, Sutton, J.M, Perugini, M.A, Soares da Costa, T.P. | | Deposit date: | 2019-06-08 | | Release date: | 2019-08-07 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Identification of two dihydrodipicolinate synthase isoforms from Pseudomonas aeruginosa that differ in allosteric regulation.
Febs J., 287, 2020
|
|
4YDX
 
 | | Crystal structure of cisplatin bound to a human copper chaperone (monomer) - new refinement | | Descriptor: | 3,3',3''-phosphanetriyltripropanoic acid, Copper transport protein ATOX1, PLATINUM (II) ION, ... | | Authors: | Shabalin, I.G, Boal, A.K, Dauter, Z, Jaskolski, M, Minor, W, Rosenzweig, A.C, Wlodawer, A. | | Deposit date: | 2015-02-23 | | Release date: | 2015-03-04 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.602 Å) | | Cite: | Crystallography and chemistry should always go together: a cautionary tale of protein complexes with cisplatin and carboplatin.
Acta Crystallogr.,Sect.D, 71, 2015
|
|
4YEA
 
 | | Crystal structure of cisplatin bound to a human copper chaperone (dimer) - new refinement | | Descriptor: | COPPER (II) ION, Copper transport protein ATOX1, SULFATE ION | | Authors: | Shabalin, I.G, Dauter, Z, Jaskolski, M, Minor, W, Wlodawer, A. | | Deposit date: | 2015-02-23 | | Release date: | 2015-03-18 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.14 Å) | | Cite: | Crystallography and chemistry should always go together: a cautionary tale of protein complexes with cisplatin and carboplatin.
Acta Crystallogr.,Sect.D, 71, 2015
|
|
4XIH
 
 | |
6EZW
 
 | | Crystal structure of a llama VHH antibody BCD090-M2 against human ErbB3 in space group C2 | | Descriptor: | VHH antibody BCD090-M2 | | Authors: | Eliseev, I.E, Yudenko, A.N, Vysochinskaya, V.V, Svirina, A.A, Evstratyeva, A.V, Drozhzhachih, M.S, Krendeleva, E.A, Vladimirova, A.K, Nemankin, T.A, Ekimova, V.M, Ulitin, A.B, Lomovskaya, M.I, Yakovlev, P.A, Moiseenko, F.V, Chakchir, O.B. | | Deposit date: | 2017-11-16 | | Release date: | 2017-11-29 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.59842849 Å) | | Cite: | Crystal structures of a llama VHH antibody BCD090-M2 targeting human ErbB3 receptor.
F1000Res, 7, 2018
|
|
4XR5
 
 | | X-ray structure of the unliganded thymidine phosphorylase from Salmonella typhimurium at 2.05 A resolution | | Descriptor: | CHLORIDE ION, DI(HYDROXYETHYL)ETHER, TRIETHYLENE GLYCOL, ... | | Authors: | Balaev, V.V, Lashkov, A.A, Gabdulkhakov, A.G, Betzel, C, Mikhailov, A.M. | | Deposit date: | 2015-01-20 | | Release date: | 2016-01-27 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Structural investigation of the thymidine phosphorylase from Salmonella typhimurium in the unliganded state and its complexes with thymidine and uridine.
Acta Crystallogr.,Sect.F, 72, 2016
|
|
6EVG
 
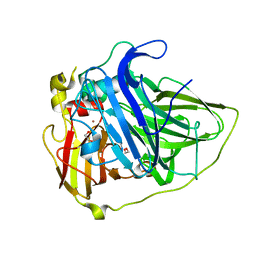 | |
6F0D
 
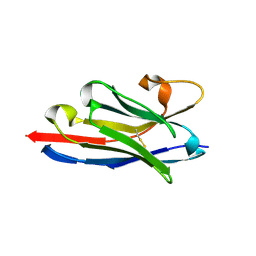 | | Crystal structure of a llama VHH antibody BCD090-M2 against human ErbB3 in space group P1 with cadmium ions | | Descriptor: | CADMIUM ION, VHH antibody BCD090-M2 | | Authors: | Eliseev, I.E, Yudenko, A.N, Vysochinskaya, V.V, Svirina, A.A, Evstratyeva, A.V, Drozhzhachih, M.S, Krendeleva, E.A, Vladimirova, A.K, Nemankin, T.A, Ekimova, V.M, Ulitin, A.B, Lomovskaya, M.I, Yakovlev, P.A, Moiseenko, F.V, Chakchir, O.B. | | Deposit date: | 2017-11-19 | | Release date: | 2017-11-29 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.90000749 Å) | | Cite: | Crystal structures of a llama VHH antibody BCD090-M2 targeting human ErbB3 receptor.
F1000Res, 7, 2018
|
|
6ISD
 
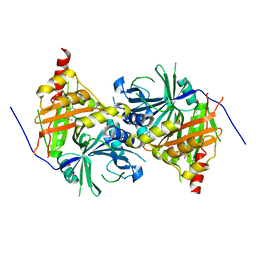 | | Crystal structure of Arabidopsis thaliana HPPD complexed with sulcotrione | | Descriptor: | 2-[2-chloro-4-(methylsulfonyl)benzoyl]cyclohexane-1,3-dione, 4-hydroxyphenylpyruvate dioxygenase, COBALT (II) ION | | Authors: | Yang, W.C, Yang, G.F. | | Deposit date: | 2018-11-16 | | Release date: | 2018-11-28 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Molecular insights into the mechanism of 4-hydroxyphenylpyruvate dioxygenase inhibition: enzyme kinetics, X-ray crystallography and computational simulations.
FEBS J., 286, 2019
|
|
1H0M
 
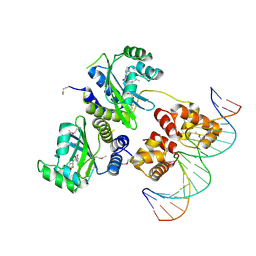 | | Three-dimensional structure of the quorum sensing protein TraR bound to its autoinducer and to its target DNA | | Descriptor: | 3-OXO-OCTANOIC ACID (2-OXO-TETRAHYDRO-FURAN-3-YL)-AMIDE, 5'-D(*AP*TP*GP*TP*GP*CP*AP*GP*AP*TP *CP*TP*GP*CP*AP*CP*AP*T)-3', Transcriptional activator protein TraR | | Authors: | Vannini, A, Volpari, C, Gargioli, C, Muraglia, E, Cortese, R, De Francesco, R, Neddermann, P, Di Marco, S. | | Deposit date: | 2002-06-25 | | Release date: | 2002-08-29 | | Last modified: | 2020-12-16 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | The Crystal Structure of the Quorum Sensing Protein Trar Bound to its Autoinducer and Target DNA
Embo J., 21, 2002
|
|
2B5Q
 
 | |
6IEV
 
 | | Crystal structure of a designed protein | | Descriptor: | Designed protein | | Authors: | Han, M, Liao, S, Chen, Q, Liu, H. | | Deposit date: | 2018-09-17 | | Release date: | 2019-09-18 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Selection and analyses of variants of a designed protein suggest importance of hydrophobicity of partially buried sidechains for protein stability at high temperatures.
Protein Sci., 28, 2019
|
|
2KB1
 
 | | NMR studies of a channel protein without membrane: structure and dynamics of water-solubilized KcsA | | Descriptor: | WSK3 | | Authors: | Ma, D, Xu, Y, Tillman, T, Tang, P, Meirovitch, E, Eckenhoff, R, Carnini, A. | | Deposit date: | 2008-11-19 | | Release date: | 2008-12-09 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | NMR studies of a channel protein without membranes: structure and dynamics of water-solubilized KcsA
Proc.Natl.Acad.Sci.USA, 105, 2008
|
|
4LFV
 
 | | Crystal structure of human FPPS in complex with YS0470 and two molecules of inorganic phosphate | | Descriptor: | CHLORIDE ION, Farnesyl pyrophosphate synthase, MAGNESIUM ION, ... | | Authors: | Park, J, Lin, Y.-S, Tsantrizos, Y.S, Berghuis, A.M. | | Deposit date: | 2013-06-27 | | Release date: | 2014-03-12 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structure of human farnesyl pyrophosphate synthase in complex with an aminopyridine bisphosphonate and two molecules of inorganic phosphate.
Acta Crystallogr F Struct Biol Commun, 70, 2014
|
|
2KDN
 
 | | Solution structure of PFE0790c, a putative bolA-like protein from the protozoan parasite Plasmodium falciparum. | | Descriptor: | Putative uncharacterized protein PFE0790c | | Authors: | Buchko, G.W, Yee, A, Semesi, A, Hui, R, Arrowsmith, C.H, Seattle Structural Genomics Center for Infectious Disease (SSGCID) | | Deposit date: | 2009-01-12 | | Release date: | 2009-01-20 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Solution-state NMR structure of the putative morphogene protein BolA (PFE0790c) from Plasmodium falciparum.
Acta Crystallogr F Struct Biol Commun, 71, 2015
|
|
2KVJ
 
 | | NMR and MD solution structure of a Gamma-Methylated PNA duplex | | Descriptor: | Gamma-Modified Peptide Nucleic Acid | | Authors: | He, W, Crawford, M.J, Rapireddy, S, Madrid, M, Gil, R.R, Ly, D.H, Achim, C. | | Deposit date: | 2010-03-15 | | Release date: | 2010-05-12 | | Last modified: | 2023-11-15 | | Method: | SOLUTION NMR | | Cite: | The structure of a gamma-modified peptide nucleic acid duplex.
Mol Biosyst, 6, 2010
|
|
2LGI
 
 | | Atomic Resolution Protein Structures using NMR Chemical Shift Tensors | | Descriptor: | Immunoglobulin G-binding protein G | | Authors: | Wylie, B.J, Sperling, L.J, Nieuwkoop, A.J, Franks, W.T, Oldfield, E, Rienstra, C.M. | | Deposit date: | 2011-07-26 | | Release date: | 2011-10-26 | | Last modified: | 2024-05-01 | | Method: | SOLID-STATE NMR | | Cite: | Ultrahigh resolution protein structures using NMR chemical shift tensors.
Proc.Natl.Acad.Sci.USA, 108, 2011
|
|
