2X8A
 
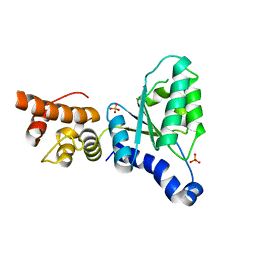 | | Human Nuclear Valosin containing protein Like (NVL), C-terminal AAA- ATPase domain | | Descriptor: | NUCLEAR VALOSIN-CONTAINING PROTEIN-LIKE, PHOSPHATE ION | | Authors: | Moche, M, Schuetz, P, Arrowsmith, C.H, Berglund, H, Bountra, C, Collins, R, Edwards, A.M, Flodin, S, Flores, A, Graslund, S, Hammarstrom, M, Johansson, I, Kallas, A, Karlberg, T, Kotenyova, T, Kraulis, P, Nordlund, P, Nyman, T, Persson, C, Sehic, A, Siponen, M.I, Svensson, L, Thorsell, A.G, Tresaugues, L, VanDenBerg, S, Wahlberg, E, Weigelt, J, Welin, M, Wisniewska, M, Schuler, H. | | Deposit date: | 2010-03-08 | | Release date: | 2010-03-16 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Human Nuclear Valosin Containing Protein Like (Nvl) , C-Terminal Aaa-ATPase Domain
To be Published
|
|
4HSE
 
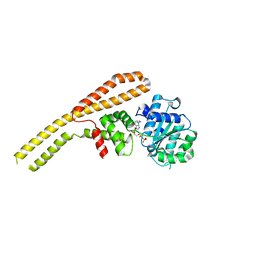 | | Crystal structure of ClpB NBD1 in complex with guanidinium chloride and ADP | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, CHLORIDE ION, Chaperone protein ClpB, ... | | Authors: | Zeymer, C, Werbeck, N.D, Schlichting, I, Reinstein, J. | | Deposit date: | 2012-10-30 | | Release date: | 2013-01-30 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | The molecular mechanism of hsp100 chaperone inhibition by the prion curing agent guanidinium chloride.
J.Biol.Chem., 288, 2013
|
|
2ZAM
 
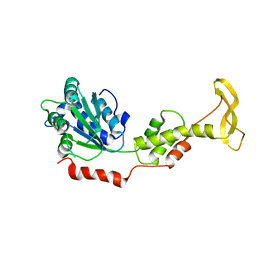 | | Crystal structure of mouse SKD1/VPS4B apo-form | | Descriptor: | Vacuolar protein sorting-associating protein 4B | | Authors: | Inoue, M, Kawasaki, M, Kamikubo, H, Kataoka, M, Kato, R, Yoshimori, T, Wakatsuki, S. | | Deposit date: | 2007-10-08 | | Release date: | 2008-10-07 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (3.5 Å) | | Cite: | Nucleotide-dependent conformational changes and assembly of the AAA ATPase SKD1/VPS4B
Traffic, 9, 2008
|
|
2ZAO
 
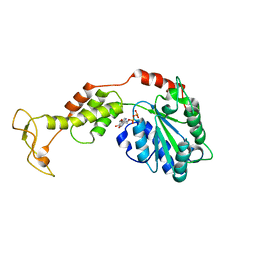 | | Crystal structure of mouse SKD1/VPS4B ADP-form | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, Vacuolar protein sorting-associating protein 4B | | Authors: | Inoue, M, Kawasaki, M, Kamikubo, H, Kataoka, M, Kato, R, Yoshimori, T, Wakatsuki, S. | | Deposit date: | 2007-10-08 | | Release date: | 2008-10-07 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Nucleotide-dependent conformational changes and assembly of the AAA ATPase SKD1/VPS4B
Traffic, 9, 2008
|
|
2ZAN
 
 | | Crystal structure of mouse SKD1/VPS4B ATP-form | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, MAGNESIUM ION, Vacuolar protein sorting-associating protein 4B | | Authors: | Inoue, M, Kawasaki, M, Kamikubo, H, Kataoka, M, Kato, R, Yoshimori, T, Wakatsuki, S. | | Deposit date: | 2007-10-08 | | Release date: | 2008-10-07 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Nucleotide-dependent conformational changes and assembly of the AAA ATPase SKD1/VPS4B
Traffic, 9, 2008
|
|
4KO8
 
 | | Structure of p97 N-D1 R155H mutant in complex with ATPgS | | Descriptor: | MAGNESIUM ION, PHOSPHOTHIOPHOSPHORIC ACID-ADENYLATE ESTER, Transitional endoplasmic reticulum ATPase | | Authors: | Xia, D, Tang, W.K. | | Deposit date: | 2013-05-11 | | Release date: | 2013-11-13 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.98 Å) | | Cite: | Altered Intersubunit Communication Is the Molecular Basis for Functional Defects of Pathogenic p97 Mutants.
J.Biol.Chem., 288, 2013
|
|
4KOD
 
 | | Structure of p97 N-D1 R155H mutant in complex with ADP | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, Transitional endoplasmic reticulum ATPase | | Authors: | Xia, D, Tang, W.K. | | Deposit date: | 2013-05-11 | | Release date: | 2013-11-13 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.96 Å) | | Cite: | Altered Intersubunit Communication Is the Molecular Basis for Functional Defects of Pathogenic p97 Mutants.
J.Biol.Chem., 288, 2013
|
|
4KLN
 
 | | Structure of p97 N-D1 A232E mutant in complex with ATPgS | | Descriptor: | MAGNESIUM ION, PHOSPHOTHIOPHOSPHORIC ACID-ADENYLATE ESTER, Transitional endoplasmic reticulum ATPase | | Authors: | Xia, D, Tang, W.K. | | Deposit date: | 2013-05-07 | | Release date: | 2013-11-13 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.62 Å) | | Cite: | Altered Intersubunit Communication Is the Molecular Basis for Functional Defects of Pathogenic p97 Mutants.
J.Biol.Chem., 288, 2013
|
|
4L15
 
 | | Crystal structure of FIGL-1 AAA domain | | Descriptor: | Fidgetin-like protein 1, TRIS(HYDROXYETHYL)AMINOMETHANE | | Authors: | Peng, W, Lin, Z, Li, W, Lu, J, Shen, Y, Wang, C. | | Deposit date: | 2013-06-02 | | Release date: | 2013-09-04 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Structural insights into the unusually strong ATPase activity of the AAA domain of the Caenorhabditis elegans fidgetin-like 1 (FIGL-1) protein.
J.Biol.Chem., 288, 2013
|
|
4L16
 
 | | Crystal structure of FIGL-1 AAA domain in complex with ADP | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, Fidgetin-like protein 1 | | Authors: | Peng, W, Lin, Z, Li, W, Lu, J, Shen, Y, Wang, C. | | Deposit date: | 2013-06-02 | | Release date: | 2013-09-04 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structural insights into the unusually strong ATPase activity of the AAA domain of the Caenorhabditis elegans fidgetin-like 1 (FIGL-1) protein.
J.Biol.Chem., 288, 2013
|
|
4LCB
 
 | | Structure of Vps4 homolog from Acidianus hospitalis | | Descriptor: | CHLORIDE ION, Cell division protein CdvC, Vps4 | | Authors: | Han, H, Hill, C.P, Whitby, F.G, Monroe, N. | | Deposit date: | 2013-06-21 | | Release date: | 2013-11-06 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.08 Å) | | Cite: | The Oligomeric State of the Active Vps4 AAA ATPase.
J.Mol.Biol., 426, 2014
|
|
4LGM
 
 | | Crystal Structure of Sulfolobus solfataricus Vps4 | | Descriptor: | CHLORIDE ION, Vps4 AAA ATPase | | Authors: | Han, H, Hill, C.P, Whitby, F.G, Monroe, N. | | Deposit date: | 2013-06-28 | | Release date: | 2013-11-06 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.711 Å) | | Cite: | The Oligomeric State of the Active Vps4 AAA ATPase.
J.Mol.Biol., 426, 2014
|
|
4WW0
 
 | | Truncated FtsH from A. aeolicus | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, ATP-dependent zinc metalloprotease FtsH, ZINC ION | | Authors: | Vostrukhina, M, Baumann, U, Schacherl, M, Bieniossek, C, Lanz, M, Baumgartner, R. | | Deposit date: | 2014-11-09 | | Release date: | 2015-05-06 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.96 Å) | | Cite: | The structure of Aquifex aeolicus FtsH in the ADP-bound state reveals a C2-symmetric hexamer.
Acta Crystallogr.,Sect.D, 71, 2015
|
|
1YYF
 
 | | Correction of X-ray Intensities from an HslV-HslU co-crystal containing lattice translocation defects | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, ATP-dependent hsl protease ATP-binding subunit hslU, ATP-dependent protease hslV | | Authors: | Wang, J, Rho, S.H, Park, H.H, Eom, S.H. | | Deposit date: | 2005-02-24 | | Release date: | 2005-07-12 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (4.16 Å) | | Cite: | Correction of X-ray intensities from an HslV-HslU co-crystal containing lattice-translocation defects.
Acta Crystallogr.,Sect.D, 61, 2005
|
|
4XGC
 
 | | Crystal structure of the eukaryotic origin recognition complex | | Descriptor: | CHLORIDE ION, Origin recognition complex subunit 1, Origin recognition complex subunit 2, ... | | Authors: | Bleichert, F, Botchan, M.R, Berger, J.M. | | Deposit date: | 2014-12-30 | | Release date: | 2015-04-01 | | Last modified: | 2019-12-04 | | Method: | X-RAY DIFFRACTION (3.5 Å) | | Cite: | Crystal structure of the eukaryotic origin recognition complex.
Nature, 519, 2015
|
|
1OFI
 
 | | Asymmetric complex between HslV and I-domain deleted HslU (H. influenzae) | | Descriptor: | 4-IODO-3-NITROPHENYL ACETYL-LEUCINYL-LEUCINYL-LEUCINYL-VINYLSULFONE, ADENOSINE-5'-DIPHOSPHATE, ATP-DEPENDENT HSL PROTEASE ATP-BINDING SUBUNIT HSLU, ... | | Authors: | Kwon, A.R, Kessler, B.M, Overkleeft, H.S, McKay, D.B. | | Deposit date: | 2003-04-14 | | Release date: | 2003-07-03 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Structure and Reactivity of an Asymmetric Complex between Hslv and I-Domain Deleted Hslu, a Prokaryotic Homolog of the Eukaryotic Proteasome
J.Mol.Biol., 330, 2003
|
|
1OFH
 
 | | Asymmetric complex between HslV and I-domain deleted HslU (H. influenzae) | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, ATP-DEPENDENT HSL PROTEASE ATP-BINDING SUBUNIT HSLU, ATP-DEPENDENT PROTEASE HSLV, ... | | Authors: | Kwon, A.R, Kessler, B.M, Overkleeft, H.S, McKay, D.B. | | Deposit date: | 2003-04-14 | | Release date: | 2003-07-03 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structure and Reactivity of an Asymmetric Complex between Hslv and I-Domain Deleted Hslu, a Prokaryotic Homolog of the Eukaryotic Proteasome
J.Mol.Biol., 330, 2003
|
|
4YPL
 
 | | Crystal structure of a hexameric LonA protease bound to three ADPs | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, Lon protease, N-[(1R)-1-(dihydroxyboranyl)-2-phenylethyl]-Nalpha-(pyrazin-2-ylcarbonyl)-L-phenylalaninamide | | Authors: | Lin, C.-C, Chang, C.-I. | | Deposit date: | 2015-03-13 | | Release date: | 2016-03-16 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (3.45 Å) | | Cite: | Structural Insights into the Allosteric Operation of the Lon AAA+ Protease
Structure, 24, 2016
|
|
4Z8X
 
 | | Truncated FtsH from A. aeolicus | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, ATP-dependent zinc metalloprotease FtsH, SULFATE ION, ... | | Authors: | Vostrukhina, M, Baumann, U, Schacherl, M, Bieniossek, C, Lanz, M, Baumgartner, R. | | Deposit date: | 2015-04-09 | | Release date: | 2015-05-06 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (3.25 Å) | | Cite: | The structure of Aquifex aeolicus FtsH in the ADP-bound state reveals a C2-symmetric hexamer.
Acta Crystallogr.,Sect.D, 71, 2015
|
|
5A5B
 
 | | Structure of the 26S proteasome-Ubp6 complex | | Descriptor: | 26S PROTEASE REGULATORY SUBUNIT 4 HOMOLOG, 26S PROTEASE REGULATORY SUBUNIT 6A, 26S PROTEASE REGULATORY SUBUNIT 6B HOMOLOG, ... | | Authors: | Aufderheide, A, Beck, F, Stengel, F, Hartwig, M, Schweitzer, A, Pfeifer, G, Goldberg, A.L, Sakata, E, Baumeister, W, Foerster, F. | | Deposit date: | 2015-06-17 | | Release date: | 2015-07-22 | | Last modified: | 2017-08-30 | | Method: | ELECTRON MICROSCOPY (9.5 Å) | | Cite: | Structural Characterization of the Interaction of Ubp6 with the 26S Proteasome.
Proc.Natl.Acad.Sci.USA, 112, 2015
|
|
2CHV
 
 | | Replication Factor C ADPNP complex | | Descriptor: | REPLICATION FACTOR C SMALL SUBUNIT | | Authors: | Seybert, A, Singleton, M.R, Cook, N, Hall, D.R, Wigley, D.B. | | Deposit date: | 2006-03-16 | | Release date: | 2006-06-06 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (4 Å) | | Cite: | Communication between Subunits within an Archaeal Clamp-Loader Complex.
Embo J., 25, 2006
|
|
2CE7
 
 | | EDTA treated | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, CELL DIVISION PROTEIN FTSH, MAGNESIUM ION, ... | | Authors: | Bieniossek, C, Baumann, U. | | Deposit date: | 2006-02-03 | | Release date: | 2006-02-09 | | Last modified: | 2018-03-07 | | Method: | X-RAY DIFFRACTION (2.44 Å) | | Cite: | The Molecular Architecture of the Metalloprotease Ftsh.
Proc.Natl.Acad.Sci.USA, 103, 2006
|
|
2CHQ
 
 | | Replication Factor C ADPNP complex | | Descriptor: | PHOSPHOAMINOPHOSPHONIC ACID-ADENYLATE ESTER, REPLICATION FACTOR C SMALL SUBUNIT | | Authors: | Seybert, A, Singleton, M.R, Cook, N, Hall, D.R, Wigley, D.B. | | Deposit date: | 2006-03-16 | | Release date: | 2006-06-06 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (3.5 Å) | | Cite: | Communication between Subunits within an Archaeal Clamp-Loader Complex.
Embo J., 25, 2006
|
|
2CEA
 
 | | CELL DIVISION PROTEIN FTSH | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, CELL DIVISION PROTEIN FTSH, MAGNESIUM ION, ... | | Authors: | Bieniossek, C, Baumann, U. | | Deposit date: | 2006-02-03 | | Release date: | 2006-02-09 | | Last modified: | 2019-07-24 | | Method: | X-RAY DIFFRACTION (2.75 Å) | | Cite: | The Molecular Architecture of the Metalloprotease Ftsh.
Proc.Natl.Acad.Sci.USA, 103, 2006
|
|
1QVR
 
 | | Crystal Structure Analysis of ClpB | | Descriptor: | ClpB protein, PHOSPHOAMINOPHOSPHONIC ACID-ADENYLATE ESTER, PLATINUM (II) ION | | Authors: | Lee, S, Sowa, M.E, Watanabe, Y, Sigler, P.B, Chiu, W, Yoshida, M, Tsai, F.T.F. | | Deposit date: | 2003-08-28 | | Release date: | 2003-10-21 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | The Structure of ClpB: A Molecular Chaperone that Rescues Proteins from an Aggregated State
Cell(Cambridge,Mass.), 115, 2003
|
|
