4WOC
 
 | | Proteinase-K Post-Surface Acoustic Waves | | Descriptor: | Proteinase K, SULFATE ION | | Authors: | French, J.B. | | Deposit date: | 2014-10-15 | | Release date: | 2015-02-18 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.601 Å) | | Cite: | Precise Manipulation and Patterning of Protein Crystals for Macromolecular Crystallography Using Surface Acoustic Waves.
Small, 11, 2015
|
|
4WOB
 
 | | Proteinase-K Pre-Surface Acoustic Wave | | Descriptor: | Proteinase K, SULFATE ION | | Authors: | French, J.B. | | Deposit date: | 2014-10-15 | | Release date: | 2015-02-18 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Precise Manipulation and Patterning of Protein Crystals for Macromolecular Crystallography Using Surface Acoustic Waves.
Small, 11, 2015
|
|
7W2A
 
 | | gliadinase Bga1903 | | Descriptor: | CALCIUM ION, Peptidase | | Authors: | Meng, M. | | Deposit date: | 2021-11-23 | | Release date: | 2022-12-07 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Crystal structure of a Burkholderia peptidase and modification of the substrate-binding site for enhanced hydrolytic activity toward gluten-derived pro-immunogenic peptides.
Int.J.Biol.Macromol., 222, 2022
|
|
5GL8
 
 | |
5I9S
 
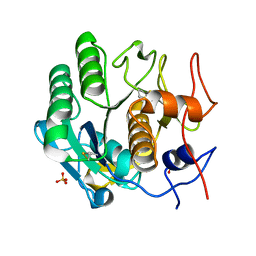 | | MicroED structure of proteinase K at 1.75 A resolution | | Descriptor: | Proteinase K, SULFATE ION | | Authors: | Hattne, J, Shi, D, de la Cruz, M.J, Reyes, F.E, Gonen, T. | | Deposit date: | 2016-02-20 | | Release date: | 2016-06-08 | | Last modified: | 2023-08-30 | | Method: | ELECTRON CRYSTALLOGRAPHY (1.75 Å) | | Cite: | Modeling truncated pixel values of faint reflections in MicroED images.
J.Appl.Crystallogr., 49, 2016
|
|
7JSY
 
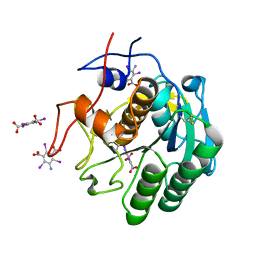 | |
7XR8
 
 | |
7Y6M
 
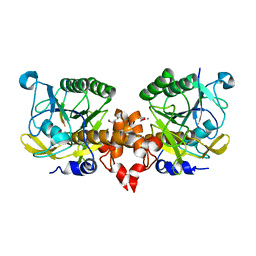 | |
4ZOQ
 
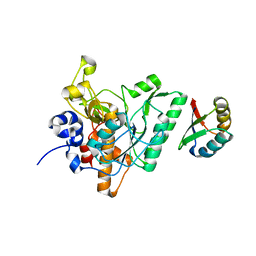 | | Crystal Structure of a Lanthipeptide Protease | | Descriptor: | Intracellular serine protease | | Authors: | Dong, S.H, Nair, S.K. | | Deposit date: | 2015-05-06 | | Release date: | 2016-03-23 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Applications of the class II lanthipeptide protease LicP for sequence-specific, traceless peptide bond cleavage.
Chem Sci, 6, 2015
|
|
4ZAR
 
 | | Crystal Structure of Proteinase K from Engyodontium albuminhibited by METHOXYSUCCINYL-ALA-ALA-PRO-PHE-CHLOROMETHYL KETONE at 1.15 A resolution | | Descriptor: | CALCIUM ION, METHOXYSUCCINYL-ALA-ALA-PRO-PHE-CHLOROMETHYL KETONE, bound form, ... | | Authors: | Sawaya, M.R, Cascio, D, Collazo, M, Bond, C, Cohen, A, DeNicola, A, Eden, K, Jain, K, Leung, C, Lubock, N, McCormick, J, Rosinski, J, Spiegelman, L, Athar, Y, Tibrewal, N, Winter, J, Solomon, S. | | Deposit date: | 2015-04-14 | | Release date: | 2015-05-06 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.15 Å) | | Cite: | Crystal Structure of Proteinase K from Engyodontium album inhibited by METHOXYSUCCINYL-ALA-ALA-PRO-PHE-CHLOROMETHYL KETONE at 1.15 A resolution
to be published
|
|
8SDK
 
 | | The MicroED structure of proteinase K crystallized by suspended drop crystallization | | Descriptor: | CALCIUM ION, Proteinase K, SULFATE ION | | Authors: | Gillman, C, Nicolas, W.J, Martynowycz, M.W, Gonen, T. | | Deposit date: | 2023-04-06 | | Release date: | 2023-05-31 | | Last modified: | 2023-07-12 | | Method: | ELECTRON CRYSTALLOGRAPHY (2.1 Å) | | Cite: | Design and implementation of suspended drop crystallization.
Iucrj, 10, 2023
|
|
8SQV
 
 | | Proteinase K Multiconformer Model at 333K | | Descriptor: | CALCIUM ION, Proteinase K, SULFATE ION | | Authors: | Du, S, Wankowicz, S, Yabukarski, F, Doukov, T, Herschlag, D, Fraser, J.S. | | Deposit date: | 2023-05-04 | | Release date: | 2023-08-09 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.22 Å) | | Cite: | Refinement of multiconformer ensemble models from multi-temperature X-ray diffraction data.
Methods Enzymol., 688, 2023
|
|
1AK9
 
 | | SUBTILISIN MUTANT 8321 | | Descriptor: | CALCIUM ION, ISOPROPYL ALCOHOL, SODIUM ION, ... | | Authors: | Whitlow, M, Howard, A.J, Wood, J.F. | | Deposit date: | 1997-05-30 | | Release date: | 1997-11-12 | | Last modified: | 2021-11-03 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Large increases in general stability for subtilisin BPN' through incremental changes in the free energy of unfolding.
Biochemistry, 28, 1989
|
|
8SPL
 
 | | Proteinase K Multiconformer Model at 343K | | Descriptor: | ALA-ALA-ALA-SER-VAL-LYS, CALCIUM ION, Proteinase K, ... | | Authors: | Du, S, Wankowicz, S, Yabukarski, F, Doukov, T, Herschlag, D, Fraser, J.S. | | Deposit date: | 2023-05-03 | | Release date: | 2023-08-09 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.21 Å) | | Cite: | Refinement of multiconformer ensemble models from multi-temperature X-ray diffraction data.
Methods Enzymol., 688, 2023
|
|
8SOV
 
 | | Proteinase K Multiconformer Model at 353K | | Descriptor: | ALA-ALA-ALA-SER-VAL-LYS, CALCIUM ION, Proteinase K, ... | | Authors: | Du, S, Wankowicz, S, Yabukarski, F, Doukov, T, Herschlag, D, Fraser, J.S. | | Deposit date: | 2023-04-30 | | Release date: | 2023-08-09 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.291 Å) | | Cite: | Refinement of multiconformer ensemble models from multi-temperature X-ray diffraction data.
Methods Enzymol., 688, 2023
|
|
8SOG
 
 | | Proteinase K Multiconformer Model at 313K | | Descriptor: | CALCIUM ION, Proteinase K, SULFATE ION | | Authors: | Du, S, Wankowicz, S, Yabukarski, F, Doukov, T, Herschlag, D, Fraser, J.S. | | Deposit date: | 2023-04-28 | | Release date: | 2023-08-09 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.13 Å) | | Cite: | Refinement of multiconformer ensemble models from multi-temperature X-ray diffraction data.
Methods Enzymol., 688, 2023
|
|
8SOU
 
 | | Proteinase K Multiconformer Model at 363K | | Descriptor: | ALA-ALA-ALA-SER-VAL-LYS, CALCIUM ION, Proteinase K, ... | | Authors: | Du, S, Wankowicz, S, Yabukarski, F, Doukov, T, Herschlag, D, Fraser, J.S. | | Deposit date: | 2023-04-30 | | Release date: | 2023-08-09 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.54 Å) | | Cite: | Refinement of multiconformer ensemble models from multi-temperature X-ray diffraction data.
Methods Enzymol., 688, 2023
|
|
1A2Q
 
 | | SUBTILISIN BPN' MUTANT 7186 | | Descriptor: | ACETONE, CALCIUM ION, SUBTILISIN BPN' | | Authors: | Gilliland, G.L, Whitlow, M, Howard, A.J. | | Deposit date: | 1998-01-08 | | Release date: | 1998-04-29 | | Last modified: | 2021-11-03 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Large increases in general stability for subtilisin BPN' through incremental changes in the free energy of unfolding.
Biochemistry, 28, 1989
|
|
1AF4
 
 | | CRYSTAL STRUCTURE OF SUBTILISIN CARLSBERG IN ANHYDROUS DIOXANE | | Descriptor: | 1,4-DIETHYLENE DIOXIDE, CALCIUM ION, SUBTILISIN CARLSBERG | | Authors: | Schmitke, J.L, Stern, L.J, Klibanov, A.M. | | Deposit date: | 1997-03-21 | | Release date: | 1997-06-16 | | Last modified: | 2023-08-02 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | The crystal structure of subtilisin Carlsberg in anhydrous dioxane and its comparison with those in water and acetonitrile.
Proc.Natl.Acad.Sci.USA, 94, 1997
|
|
1AH2
 
 | | SERINE PROTEASE PB92 FROM BACILLUS ALCALOPHILUS, NMR, 18 STRUCTURES | | Descriptor: | SERINE PROTEASE PB92 | | Authors: | Boelens, R, Schipper, D, Martin, J.R, Karimi-Nejad, Y, Mulder, F, Zwan, J.V.D, Mariani, M. | | Deposit date: | 1997-04-11 | | Release date: | 1998-04-15 | | Last modified: | 2022-02-16 | | Method: | SOLUTION NMR | | Cite: | The solution structure of serine protease PB92 from Bacillus alcalophilus presents a rigid fold with a flexible substrate-binding site.
Structure, 5, 1997
|
|
1AV7
 
 | | SUBTILISIN CARLSBERG L-NAPHTHYL-1-ACETAMIDO BORONIC ACID INHIBITOR COMPLEX | | Descriptor: | SODIUM ION, SUBTILISIN CARLSBERG, TYPE VIII | | Authors: | Stoll, V.S, Eger, B.T, Hynes, R.C, Martichonok, V, Jones, J.B, Pai, E.F. | | Deposit date: | 1997-09-29 | | Release date: | 1998-04-01 | | Last modified: | 2023-08-02 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Differences in binding modes of enantiomers of 1-acetamido boronic acid based protease inhibitors: crystal structures of gamma-chymotrypsin and subtilisin Carlsberg complexes.
Biochemistry, 37, 1998
|
|
1AQN
 
 | | SUBTILISIN MUTANT 8324 | | Descriptor: | CALCIUM ION, ISOPROPYL ALCOHOL, SUBTILISIN 8324, ... | | Authors: | Whitlow, M, Howard, A.J, Wood, J.F. | | Deposit date: | 1997-07-31 | | Release date: | 1998-01-14 | | Last modified: | 2021-11-03 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Large increases in general stabilityfor subtilisin BPN' through incremental changes in the free energy of unfolding
To be published
|
|
1AVT
 
 | | SUBTILISIN CARLSBERG D-PARA-CHLOROPHENYL-1-ACETAMIDO BORONIC ACID INHIBITOR COMPLEX | | Descriptor: | SODIUM ION, SUBTILISIN CARLSBERG, TYPE VIII | | Authors: | Stoll, V.S, Eger, B.T, Hynes, R.C, Martichonok, V, Jones, J.B, Pai, E.F. | | Deposit date: | 1997-09-19 | | Release date: | 1998-03-25 | | Last modified: | 2023-08-02 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Differences in binding modes of enantiomers of 1-acetamido boronic acid based protease inhibitors: crystal structures of gamma-chymotrypsin and subtilisin Carlsberg complexes.
Biochemistry, 37, 1998
|
|
6CL7
 
 | | 1.71 A MicroED structure of proteinase K at 0.86 e- / A^2 | | Descriptor: | Proteinase K | | Authors: | Hattne, J, Shi, D, Glynn, C, Zee, C.-T, Gallagher-Jones, M, Martynowycz, M.W, Rodriguez, J.A, Gonen, T. | | Deposit date: | 2018-03-02 | | Release date: | 2018-05-16 | | Last modified: | 2023-10-04 | | Method: | ELECTRON CRYSTALLOGRAPHY (1.71 Å) | | Cite: | Analysis of Global and Site-Specific Radiation Damage in Cryo-EM.
Structure, 26, 2018
|
|
5K7S
 
 | | MicroED structure of proteinase K at 1.6 A resolution | | Descriptor: | CALCIUM ION, Proteinase K | | Authors: | de la Cruz, M.J, Hattne, J, Shi, D, Seidler, P, Rodriguez, J, Reyes, F.E, Sawaya, M.R, Cascio, D, Eisenberg, D, Gonen, T. | | Deposit date: | 2016-05-26 | | Release date: | 2017-04-05 | | Last modified: | 2018-08-22 | | Method: | ELECTRON CRYSTALLOGRAPHY (1.6 Å) | | Cite: | Atomic-resolution structures from fragmented protein crystals with the cryoEM method MicroED.
Nat. Methods, 14, 2017
|
|
