3RHG
 
 | | Crystal structure of amidohydrolase pmi1525 (target efi-500319) from proteus mirabilis hi4320 | | Descriptor: | BENZOIC ACID, CACODYLATE ION, Putative phophotriesterase, ... | | Authors: | Patskovsky, Y, Hillerich, B, Seidel, R.D, Zencheck, W.D, Toro, R, Imker, H.J, Raushel, F.M, Gerlt, J.A, Almo, S.C, Enzyme Function Initiative (EFI) | | Deposit date: | 2011-04-11 | | Release date: | 2011-04-27 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.53 Å) | | Cite: | Crystal Structure of Amidohydrolase Pmi1525 from Proteus Mirabilis Hi4320
To be Published
|
|
4LEF
 
 | | Crystal structure of PHOSPHOTRIESTERASE HOMOLOGY PROTEIN FROM ESCHERICHIA COLI complexed with phosphate in active site | | Descriptor: | PHOSPHATE ION, Phosphotriesterase homology protein, ZINC ION, ... | | Authors: | Fedorov, A.A, Fedorov, E.V, Xiang, D.F, Raushel, F.M, Almo, S.C. | | Deposit date: | 2013-06-25 | | Release date: | 2014-06-25 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.842 Å) | | Cite: | Crystal structure of PHOSPHOTRIESTERASE HOMOLOGY PROTEIN FROM ESCHERICHIA COLI complexed with phosphate in active site
To be Published
|
|
5CH9
 
 | | Gkap mutant B12 | | Descriptor: | COBALT (II) ION, Phosphotriesterase | | Authors: | Chen, L.-Q. | | Deposit date: | 2015-07-10 | | Release date: | 2015-09-09 | | Last modified: | 2019-11-20 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Active site loop architecture enhance the promiscuous PTE activity in lactonase from Geobacillus kaustophilus HTA426
To Be Published
|
|
3WML
 
 | | Structure of phosphotriesterase mutant (S308L/Y309A) from Agrobacterium radiobacter | | Descriptor: | 1,2-ETHANEDIOL, COBALT (II) ION, FE (II) ION, ... | | Authors: | Jackson, C.J, Carr, P.D, Sugrue, E. | | Deposit date: | 2013-11-21 | | Release date: | 2014-09-24 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (1.99 Å) | | Cite: | A 5000-fold increase in the specificity of a bacterial phosphotriesterase for malathion through combinatorial active site mutagenesis
Plos One, 9, 2014
|
|
6BHL
 
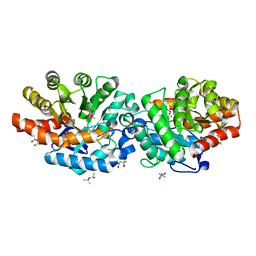 | | Phosphotriesterase variant S5deltaL7 | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, CACODYLATE ION, Phosphotriesterase, ... | | Authors: | Miton, C.M, Campbell, E.C, Jackson, C.J, Tokuriki, N. | | Deposit date: | 2017-10-30 | | Release date: | 2019-01-23 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Phosphotriesterase variant S5deltaL7
To Be Published
|
|
1BF6
 
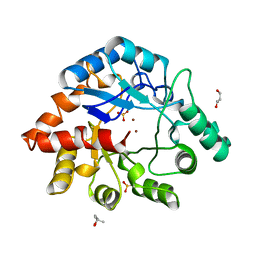 | | PHOSPHOTRIESTERASE HOMOLOGY PROTEIN FROM ESCHERICHIA COLI | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, GLYCEROL, PHOSPHOTRIESTERASE HOMOLOGY PROTEIN, ... | | Authors: | Buchbinder, J.L, Stephenson, R.C, Scanlan, T.S, Fletterick, R.J. | | Deposit date: | 1998-05-27 | | Release date: | 1999-06-08 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Biochemical characterization and crystallographic structure of an Escherichia coli protein from the phosphotriesterase gene family.
Biochemistry, 37, 1998
|
|
7P85
 
 | |
3MSR
 
 | |
4ZSU
 
 | |
4ZST
 
 | |
4XD6
 
 | | Phosphotriesterase Variant E2a | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, CACODYLATE ION, Phosphotriesterase variant PTE-E2, ... | | Authors: | Jackson, C.J, Campbell, E, Kaltenbach, M, Tokuriki, N. | | Deposit date: | 2014-12-19 | | Release date: | 2015-12-23 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | The role of protein dynamics in the evolution of new enzyme function.
Nat.Chem.Biol., 12, 2016
|
|
4XD5
 
 | | Phosphotriesterase variant R2 | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, CACODYLATE ION, Phosphotriesterase variant PTE-R2, ... | | Authors: | Campbell, E, Kaltenbach, M, Tokuriki, N, Jackson, C.J. | | Deposit date: | 2014-12-19 | | Release date: | 2015-12-23 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | The role of protein dynamics in the evolution of new enzyme function.
Nat.Chem.Biol., 12, 2016
|
|
2D2H
 
 | | OpdA from Agrobacterium radiobacter with bound inhibitor trimethyl phosphate at 1.8 A resolution | | Descriptor: | COBALT (II) ION, TRIMETHYL PHOSPHATE, phosphotriesterase | | Authors: | Jackson, C, Kim, H.K, Carr, P.D, Liu, J.W, Ollis, D.L. | | Deposit date: | 2005-09-09 | | Release date: | 2005-09-20 | | Last modified: | 2015-08-19 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | The structure of an enzyme-product complex reveals the critical role of a terminal hydroxide nucleophile in the bacterial phosphotriesterase mechanism
Biochim.Biophys.Acta, 1752, 2005
|
|
4XAF
 
 | | Cycles of destabilization and repair underlie evolutionary transitions in enzymes | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, CACODYLATE ION, Phosphotriesterase variant PTE-R1, ... | | Authors: | Jackson, C.J, Campbell, E, Kaltenbach, M, Tokuriki, N. | | Deposit date: | 2014-12-14 | | Release date: | 2015-12-16 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.66 Å) | | Cite: | The role of protein dynamics in the evolution of new enzyme function.
Nat.Chem.Biol., 12, 2016
|
|
1DPM
 
 | | THREE-DIMENSIONAL STRUCTURE OF THE ZINC-CONTAINING PHOSPHOTRIESTERASE WITH BOUND SUBSTRATE ANALOG DIETHYL 4-METHYLBENZYLPHOSPHONATE | | Descriptor: | DIETHYL 4-METHYLBENZYLPHOSPHONATE, FORMIC ACID, PHOSPHOTRIESTERASE, ... | | Authors: | Vanhooke, J.L, Benning, M.M, Raushel, F.M, Holden, H.M. | | Deposit date: | 1996-02-13 | | Release date: | 1997-08-20 | | Last modified: | 2024-06-05 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Three-dimensional structure of the zinc-containing phosphotriesterase with the bound substrate analog diethyl 4-methylbenzylphosphonate.
Biochemistry, 35, 1996
|
|
1EYW
 
 | | THREE-DIMENSIONAL STRUCTURE OF THE ZINC-CONTAINING PHOSPHOTRIESTERASE WITH BOUND SUBSTRATE ANALOG TRIETHYLPHOSPHATE | | Descriptor: | 2-PHENYL-ETHANOL, PHOSPHOTRIESTERASE, TRIETHYL PHOSPHATE, ... | | Authors: | Holden, H.M, Benning, M.M, Raushel, F.M, Hong, S.-B. | | Deposit date: | 2000-05-09 | | Release date: | 2000-12-20 | | Last modified: | 2017-10-04 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | The binding of substrate analogs to phosphotriesterase.
J.Biol.Chem., 275, 2000
|
|
6JST
 
 | |
6JSS
 
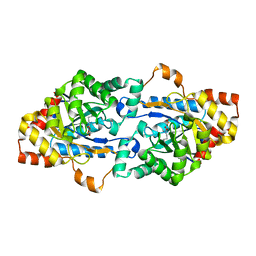 | | Structure of Geobacillus kaustophilus lactonase, Y99P mutant | | Descriptor: | FE (III) ION, HYDROXIDE ION, Phosphotriesterase, ... | | Authors: | Xue, B, Yew, W.S. | | Deposit date: | 2019-04-08 | | Release date: | 2020-04-08 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.16 Å) | | Cite: | Directed Computational Evolution of Quorum-Quenching Lactonases from the Amidohydrolase Superfamily.
Structure, 28, 2020
|
|
6JSU
 
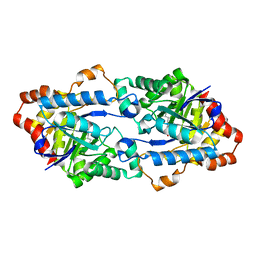 | |
1EZ2
 
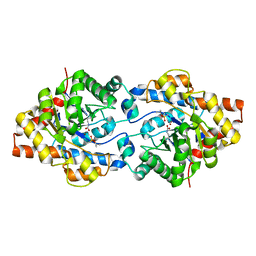 | | THREE-DIMENSIONAL STRUCTURE OF THE ZINC-CONTAINING PHOSPHOTRIESTERASE WITH BOUND SUBSTRATE ANALOG DIISOPROPYLMETHYL PHOSPHONATE. | | Descriptor: | METHYLPHOSPHONIC ACID DIISOPROPYL ESTER, PHOSPHOTRIESTERASE, ZINC ION | | Authors: | Holden, H.M, Benning, M.M, Raushel, F.M, Hong, S.-B. | | Deposit date: | 2000-05-09 | | Release date: | 2000-12-20 | | Last modified: | 2017-10-04 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | The binding of substrate analogs to phosphotriesterase.
J.Biol.Chem., 275, 2000
|
|
2D2J
 
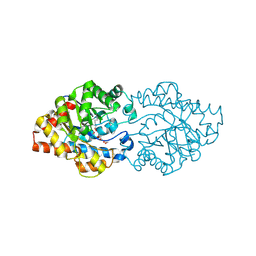 | | OpdA from Agrobacterium radiobacter without inhibitor/product present at 1.75 A resolution | | Descriptor: | 1,2-ETHANEDIOL, COBALT (II) ION, phosphotriesterase | | Authors: | Jackson, C, Kim, H.K, Carr, P.D, Liu, J.W, Ollis, D.L. | | Deposit date: | 2005-09-09 | | Release date: | 2005-09-20 | | Last modified: | 2015-08-19 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | The structure of an enzyme-product complex reveals the critical role of a terminal hydroxide nucleophile in the bacterial phosphotriesterase mechanism
Biochim.Biophys.Acta, 1752, 2005
|
|
2D2G
 
 | | OpdA from Agrobacterium radiobacter with bound product dimethylthiophosphate | | Descriptor: | COBALT (II) ION, O,O-DIMETHYL HYDROGEN THIOPHOSPHATE, phosphotriesterase | | Authors: | Jackson, C, Kim, H.K, Carr, P.D, Liu, J.W, Ollis, D.L. | | Deposit date: | 2005-09-08 | | Release date: | 2005-09-20 | | Last modified: | 2015-08-19 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | The structure of an enzyme-product complex reveals the critical role of a terminal hydroxide nucleophile in the bacterial phosphotriesterase mechanism
Biochim.Biophys.Acta, 1752, 2005
|
|
8UQW
 
 | | Round 18 Arylesterase Variant of Apo-Phosphotriesterase Measured at 13 keV | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, CHLORIDE ION, Phosphotriesterase variant PTE-R18 | | Authors: | Breeze, C.W, Frkic, R.L, Campbell, E.C, Jackson, C.J. | | Deposit date: | 2023-10-25 | | Release date: | 2024-04-03 | | Last modified: | 2024-04-10 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Mononuclear binding and catalytic activity of europium(III) and gadolinium(III) at the active site of the model metalloenzyme phosphotriesterase.
Acta Crystallogr D Struct Biol, 80, 2024
|
|
8UQZ
 
 | | Round 18 Arylesterase Variant of Phosphotriesterase Bound to Gadolinium(III) Measured at 9.5 keV | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, CHLORIDE ION, GADOLINIUM ION, ... | | Authors: | Breeze, C.W, Frkic, R.L, Campbell, E.C, Jackson, C.J. | | Deposit date: | 2023-10-25 | | Release date: | 2024-04-03 | | Last modified: | 2024-04-10 | | Method: | X-RAY DIFFRACTION (1.61 Å) | | Cite: | Mononuclear binding and catalytic activity of europium(III) and gadolinium(III) at the active site of the model metalloenzyme phosphotriesterase.
Acta Crystallogr D Struct Biol, 80, 2024
|
|
8UQX
 
 | | Round 18 Arylesterase Variant of Apo-Phosphotriesterase Measured at 9.5 keV | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, CHLORIDE ION, Phosphotriesterase variant PTE-R18 | | Authors: | Breeze, C.W, Frkic, R.L, Campbell, E.C, Jackson, C.J. | | Deposit date: | 2023-10-25 | | Release date: | 2024-04-03 | | Last modified: | 2024-04-10 | | Method: | X-RAY DIFFRACTION (1.52 Å) | | Cite: | Mononuclear binding and catalytic activity of europium(III) and gadolinium(III) at the active site of the model metalloenzyme phosphotriesterase.
Acta Crystallogr D Struct Biol, 80, 2024
|
|
