8XWX
 
 | | Crystal structure of FIS1-BAP31 complex from human | | Descriptor: | B-cell receptor-associated protein 31, BETA-MERCAPTOETHANOL, Mitochondrial fission 1 protein, ... | | Authors: | Nguyen, M.D, Bong, S.M, Lee, B.I. | | Deposit date: | 2024-01-17 | | Release date: | 2024-05-15 | | Last modified: | 2025-08-13 | | Method: | X-RAY DIFFRACTION (2.69 Å) | | Cite: | Crystal structure of Fis1 and Bap31 provides information on protein-protein interactions at mitochondria-associated ER membranes
Commun Biol, 8, 2025
|
|
8XXG
 
 | | Crystal structure of human 8-oxoguanine glycosylase K249H mutant bound to the reaction intermediate derived from the crystal soaked into the solution at pH 4.0 under 277 K for 2.5 hours | | Descriptor: | DNA (5'-D(*AP*GP*CP*GP*TP*CP*CP*A)-3'), DNA (5'-D(*GP*GP*TP*AP*GP*AP*CP*CP*TP*GP*GP*AP*CP*GP*C)-3'), DNA (5'-D(P*GP*TP*CP*TP*AP*CP*C)-3'), ... | | Authors: | Unno, M, Koga, M, Minowa, N, Tanaka, Y. | | Deposit date: | 2024-01-18 | | Release date: | 2025-07-23 | | Last modified: | 2025-08-13 | | Method: | X-RAY DIFFRACTION (1.82 Å) | | Cite: | Capturing a glycosylase reaction intermediate in DNA repair by freeze-trapping of a pH-responsive hOGG1 mutant.
Nucleic Acids Res., 53, 2025
|
|
8XXK
 
 | | Crystal structure of human 8-oxoguanine glycosylase K249H mutant bound to the reaction intermediate derived from the crystal soaked into the solution at pH 4.0 under 298 K for 3 weeks | | Descriptor: | DNA (5'-D(*AP*GP*CP*GP*TP*CP*CP*A)-3'), DNA (5'-D(*GP*GP*TP*AP*GP*AP*CP*CP*TP*GP*GP*AP*CP*GP*C)-3'), DNA (5'-D(P*GP*TP*CP*TP*AP*CP*C)-3'), ... | | Authors: | Unno, M, Koga, M, Minowa, N, Komuro, S, Tanaka, Y. | | Deposit date: | 2024-01-18 | | Release date: | 2025-07-23 | | Last modified: | 2025-08-13 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Capturing a glycosylase reaction intermediate in DNA repair by freeze-trapping of a pH-responsive hOGG1 mutant.
Nucleic Acids Res., 53, 2025
|
|
8Y1K
 
 | | The cryo-EM structure of TdpAB in complex with AMPPNP and PT-DNA | | Descriptor: | DNA (5'-D(P*GP*CP*CP*CP*TP*TP*TP*TP*GP*CP*AP*A)-3'), DNA (5'-D(P*TP*TP*GP*CP*AP*AP*AP*AP*GP*GP*GP*C)-3'), PHOSPHOAMINOPHOSPHONIC ACID-ADENYLATE ESTER, ... | | Authors: | Chen, T.A, Qian, T. | | Deposit date: | 2024-01-25 | | Release date: | 2025-01-29 | | Last modified: | 2025-08-13 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | A DNA phosphorothioation pathway via adenylated intermediate modulates Tdp machinery.
Nat.Chem.Biol., 21, 2025
|
|
8Y1R
 
 | | in situ room temperature Laue crystallography | | Descriptor: | Lysozyme C | | Authors: | Wang, Z.J, Wang, S.S, Pan, Q.Y, Yu, L, Su, Z.H, Yang, T.Y, Wang, Y.Z, Zhang, W.Z, Hao, Q, Gao, X.Y. | | Deposit date: | 2024-01-25 | | Release date: | 2024-02-07 | | Last modified: | 2025-08-13 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Laueprocess: a software package for processing Laue diffraction data
J.Appl.Crystallogr., 58, 2025
|
|
8Y1S
 
 | | in situ room temperature Laue crystallography | | Descriptor: | Lysozyme C | | Authors: | Wang, Z.J, Wang, S.S, Pan, Q.Y, Yu, L, Su, Z.H, Yang, T.Y, Wang, Y.Z, Zhang, W.Z, Hao, Q, Gao, X.Y. | | Deposit date: | 2024-01-25 | | Release date: | 2024-12-04 | | Last modified: | 2025-08-13 | | Method: | X-RAY DIFFRACTION (1.52 Å) | | Cite: | Laueprocess: a software package for processing Laue diffraction data.
J.Appl.Crystallogr., 58, 2025
|
|
8YRM
 
 | | Iota toxin Ib pore serine-clamp mutant | | Descriptor: | CALCIUM ION, Iota toxin component Ib | | Authors: | Ninomiya, Y, Yoshida, T, Yamada, T, Kishikawa, J, Tsuge, H. | | Deposit date: | 2024-03-21 | | Release date: | 2025-03-26 | | Last modified: | 2025-08-13 | | Method: | ELECTRON MICROSCOPY (2.36 Å) | | Cite: | Serine clamp of Clostridium perfringens binary toxin BECb (CPILEb)-pore confers cytotoxicity and enterotoxicity.
Commun Biol, 8, 2025
|
|
8ZKW
 
 | | Cryo-EM structure of the Drosophila INDY (apo-asymmetric, pH 6) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, PHOSPHATIDYLETHANOLAMINE, Protein I'm not dead yet | | Authors: | Kim, S, Park, J.G, Choi, S.H, Kim, J.W, Jin, M.S. | | Deposit date: | 2024-05-17 | | Release date: | 2025-01-29 | | Last modified: | 2025-08-13 | | Method: | ELECTRON MICROSCOPY (2.77 Å) | | Cite: | Cryo-EM structures reveal the H + /citrate symport mechanism of Drosophila INDY.
Life Sci Alliance, 8, 2025
|
|
8ZKZ
 
 | | Cryo-EM structure of the Drosophila INDY (apo-asymmetric, pH8) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, PHOSPHATIDYLETHANOLAMINE, Protein I'm not dead yet | | Authors: | Kim, S, Park, J.G, Choi, S.H, Kim, J.W, Jin, M.S. | | Deposit date: | 2024-05-17 | | Release date: | 2025-01-29 | | Last modified: | 2025-08-13 | | Method: | ELECTRON MICROSCOPY (2.7 Å) | | Cite: | Cryo-EM structures reveal the H + /citrate symport mechanism of Drosophila INDY.
Life Sci Alliance, 8, 2025
|
|
8ZL1
 
 | | Cryo-EM structure of the Drosophila INDY (apo outward-open, pH 6) | | Descriptor: | PHOSPHATIDYLETHANOLAMINE, Protein I'm not dead yet | | Authors: | Kim, S, Park, J.G, Choi, S.H, Kim, J.W, Jin, M.S. | | Deposit date: | 2024-05-17 | | Release date: | 2025-01-29 | | Last modified: | 2025-08-13 | | Method: | ELECTRON MICROSCOPY (2.86 Å) | | Cite: | Cryo-EM structures reveal the H + /citrate symport mechanism of Drosophila INDY.
Life Sci Alliance, 8, 2025
|
|
8ZL2
 
 | | Cryo-EM structure of the Drosophila INDY (DIDS-bound asymmetric, pH 6) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 4,4'-Diisothiocyano-2,2'-stilbenedisulfonic acid, PHOSPHATIDYLETHANOLAMINE, ... | | Authors: | Kim, S, Park, J.G, Choi, S.H, Kim, J.W, Jin, M.S. | | Deposit date: | 2024-05-17 | | Release date: | 2025-01-29 | | Last modified: | 2025-08-13 | | Method: | ELECTRON MICROSCOPY (2.8 Å) | | Cite: | Cryo-EM structures reveal the H + /citrate symport mechanism of Drosophila INDY.
Life Sci Alliance, 8, 2025
|
|
8ZL3
 
 | | Cryo-EM structure of the Drosophila INDY (DIDS-bound outward-open, pH 6) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 4,4'-Diisothiocyano-2,2'-stilbenedisulfonic acid, PHOSPHATIDYLETHANOLAMINE, ... | | Authors: | Kim, S, Park, J.G, Choi, S.H, Kim, J.W, Jin, M.S. | | Deposit date: | 2024-05-17 | | Release date: | 2025-01-29 | | Last modified: | 2025-08-13 | | Method: | ELECTRON MICROSCOPY (2.9 Å) | | Cite: | Cryo-EM structures reveal the H + /citrate symport mechanism of Drosophila INDY.
Life Sci Alliance, 8, 2025
|
|
8ZL4
 
 | | Cryo-EM structure of the Drosophila INDY (citrate-bound inward-occluded, pH 6) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, CITRATE ANION, PHOSPHATIDYLETHANOLAMINE, ... | | Authors: | Kim, S, Park, J.G, Choi, S.H, Kim, J.W, Jin, M.S. | | Deposit date: | 2024-05-17 | | Release date: | 2025-01-29 | | Last modified: | 2025-08-13 | | Method: | ELECTRON MICROSCOPY (3.65 Å) | | Cite: | Cryo-EM structures reveal the H + /citrate symport mechanism of Drosophila INDY.
Life Sci Alliance, 8, 2025
|
|
8ZL6
 
 | | Cryo-EM structure of the Drosophila INDY (apo inward-open, pH 8) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, PHOSPHATIDYLETHANOLAMINE, Protein I'm not dead yet | | Authors: | Kim, S, Park, J.G, Choi, S.H, Kim, J.W, Jin, M.S. | | Deposit date: | 2024-05-17 | | Release date: | 2025-01-29 | | Last modified: | 2025-08-13 | | Method: | ELECTRON MICROSCOPY (2.8 Å) | | Cite: | Cryo-EM structures reveal the H + /citrate symport mechanism of Drosophila INDY.
Life Sci Alliance, 8, 2025
|
|
8ZTV
 
 | | 70S ribosome arrested by PepNL with RF2 | | Descriptor: | 1,4-DIAMINOBUTANE, 16S rRNA, 23S rRNA, ... | | Authors: | Ando, Y, Kobo, A, Nureki, O, Taguchi, H, Itoh, Y, Chadani, Y. | | Deposit date: | 2024-06-07 | | Release date: | 2025-03-19 | | Last modified: | 2025-08-13 | | Method: | ELECTRON MICROSCOPY (2.9 Å) | | Cite: | A mini-hairpin shaped nascent peptide blocks translation termination by a distinct mechanism.
Nat Commun, 16, 2025
|
|
8ZYK
 
 | | Crystal structure of hemagglutinin from HN/4-10 H3N8 influenza virus S228 mutant | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Hemagglutinin, ... | | Authors: | Hao, T.J, Chai, Y, Song, H, Gao, G.F. | | Deposit date: | 2024-06-18 | | Release date: | 2025-02-19 | | Last modified: | 2025-08-13 | | Method: | X-RAY DIFFRACTION (3.01 Å) | | Cite: | Structural basis of receptor-binding adaptation of human-infecting H3N8 influenza A virus.
J.Virol., 99, 2025
|
|
9AUD
 
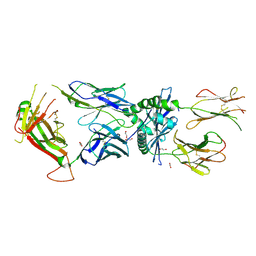 | | Immune receptor complex | | Descriptor: | 1,2-ETHANEDIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose, H-2 class II histocompatibility antigen, ... | | Authors: | Chaurasia, P, Littler, D.R, La Gruta, N, Rossjohn, J. | | Deposit date: | 2024-02-29 | | Release date: | 2025-02-05 | | Last modified: | 2025-08-13 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | LCK-co-receptor association ensures T cell lineage fidelity and maximizes epitope-specific TCR diversity.
Sci Immunol, 10, 2025
|
|
9AZI
 
 | |
9B0X
 
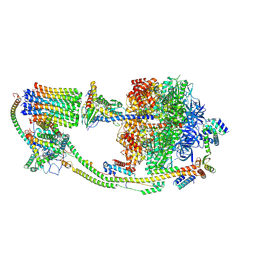 | |
9B3J
 
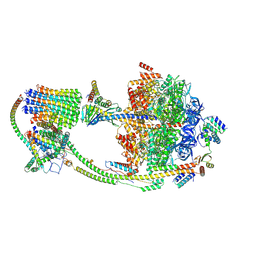 | |
9BBO
 
 | |
9BI5
 
 | | Apo form Mre11-Rad50 complex | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, DNA repair protein RAD50, Double-strand break repair protein MRE11, ... | | Authors: | Yu, Y, Patel, D.J. | | Deposit date: | 2024-04-22 | | Release date: | 2025-08-06 | | Last modified: | 2025-08-13 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | cryo EM structure of Apo form Mre11-Rad50 complex
To Be Published
|
|
9BPG
 
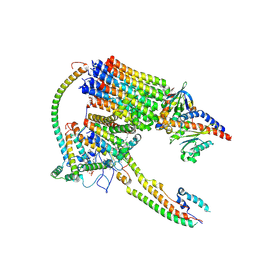 | | Artemia franciscana ATP synthase FO domain, state 1, pH 7.0 | | Descriptor: | ATP synthase protein 8, ATP synthase subunit 6.8PL, ATP synthase subunit a, ... | | Authors: | Mnatsakanyan, N, Mello, J.F.R. | | Deposit date: | 2024-05-07 | | Release date: | 2025-03-26 | | Last modified: | 2025-08-13 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Cryo-EM structure of the brine shrimp mitochondrial ATP synthase suggests an inactivation mechanism for the ATP synthase leak channel.
Cell Death Differ., 32, 2025
|
|
9BPN
 
 | |
9BU4
 
 | | Crystal structure of an MKP5 mutant, Y435W, in complex with an allosteric inhibitor | | Descriptor: | 3,3-dimethyl-1-{[9-(methylsulfanyl)-5,6-dihydrothieno[3,4-h]quinazolin-2-yl]sulfanyl}butan-2-one, Dual specificity protein phosphatase 10 | | Authors: | Manjula, R, Bennett, A.M, Lolis, E. | | Deposit date: | 2024-05-16 | | Release date: | 2025-05-28 | | Last modified: | 2025-08-13 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Dynamic and structural insights into allosteric regulation on MKP5 a dual-specificity phosphatase.
Nat Commun, 16, 2025
|
|
