2YZE
 
 | | Crystal structure of uricase from Arthrobacter globiformis | | Descriptor: | (dihydroxyboranyloxy-hydroxy-boranyl)oxylithium, Uricase | | Authors: | Juan, E.C.M, Hossain, M.T, Hoque, M.M, Suzuki, K, Sekiguchi, T, Takenaka, A. | | Deposit date: | 2007-05-05 | | Release date: | 2008-05-06 | | Last modified: | 2025-08-06 | | Method: | X-RAY DIFFRACTION (1.99 Å) | | Cite: | Structures of Arthrobacter globiformis urate oxidase-ligand complexes.
Acta Crystallogr.,Sect.D, D64, 2008
|
|
2Z6R
 
 | | Crystal structure of Lys49 to Arg mutant of Diphthine synthase | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, GLYCEROL, S-ADENOSYL-L-HOMOCYSTEINE, ... | | Authors: | Mizutani, H, Matsuura, Y, Krishna Swamy, B.S, Simanshu, D.K, Murthy, M.R.N, Kunishima, N. | | Deposit date: | 2007-08-08 | | Release date: | 2007-08-28 | | Last modified: | 2025-08-06 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Systematic study on crystal-contact engineering of diphthine synthase: influence of mutations at crystal-packing regions on X-ray diffraction quality.
Acta Crystallogr.,Sect.D, 64, 2008
|
|
3D4T
 
 | |
3DML
 
 | |
4END
 
 | | Crystal structure of anti-HIV actinohivin in complex with alpha-1,2-mannobiose (P 2 21 21 form) | | Descriptor: | ACETONITRILE, Actinohivin, alpha-D-mannopyranose-(1-2)-alpha-D-mannopyranose | | Authors: | Hoque, M.M, Suzuki, K, Tsunoda, M, Jiang, J, Zhang, F, Takahashi, A, Naomi, O, Zhang, X, Sekiguchi, T, Tanaka, H, Omura, S, Takenaka, A. | | Deposit date: | 2012-04-13 | | Release date: | 2013-07-17 | | Last modified: | 2025-08-06 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Peculiarity in crystal packing of anti-HIV lectin actinohivin in complex with alpha (1-2)mannobiose.
Acta Crystallogr.,Sect.D, 69, 2013
|
|
4G1R
 
 | | Crystal structure of anti-HIV actinohivin in complex with alphs-1,2-mannobiose (Form II) | | Descriptor: | Actinohivin, alpha-D-mannopyranose-(1-2)-alpha-D-mannopyranose | | Authors: | Hoque, M.M, Suzuki, K, Tsunoda, M, Jiang, J, Zhang, F, Takahashi, A, Naomi, O, Zhang, X, Sekiguchi, T, Tanaka, H, Omura, S, Takenaka, A. | | Deposit date: | 2012-07-11 | | Release date: | 2013-07-17 | | Last modified: | 2025-08-06 | | Method: | X-RAY DIFFRACTION (1.57 Å) | | Cite: | Peculiarity in crystal packing of anti-HIV lectin actinohivin in complex with alpha (1-2)mannobiose.
Acta Crystallogr.,Sect.D, 69, 2013
|
|
5LHB
 
 | | POLYADPRIBOSYL GLYCOSIDASE IN COMPLEX WITH PDD00017262 | | Descriptor: | 1-(cyclopropylmethyl)-6-[[(1-methylcyclopropyl)amino]-bis(oxidanyl)-$l^{4}-sulfanyl]-3-[(2-methyl-1,3-thiazol-5-yl)methyl]quinazoline-2,4-dione, DIMETHYL SULFOXIDE, Poly(ADP-ribose) glycohydrolase, ... | | Authors: | Tucker, J, Barkauskaite, E. | | Deposit date: | 2016-07-10 | | Release date: | 2016-10-12 | | Last modified: | 2025-08-06 | | Method: | X-RAY DIFFRACTION (2.23 Å) | | Cite: | First-in-Class Chemical Probes against Poly(ADP-ribose) Glycohydrolase (PARG) Inhibit DNA Repair with Differential Pharmacology to Olaparib.
ACS Chem. Biol., 11, 2016
|
|
5SDX
 
 | | CRYSTAL STRUCTURE OF HUMAN PHOSPHODIESTERASE 10 IN COMPLEX WITH c1(cccc(n1)CCc2nc(cn2C)c3ccccc3)N4CCOCC4, micromolar IC50=3.142777 | | Descriptor: | 4-{6-[(E)-2-(1-methyl-4-phenyl-1H-imidazol-2-yl)ethen-1-yl]pyridin-2-yl}morpholine, MAGNESIUM ION, ZINC ION, ... | | Authors: | Joseph, C, Groebke-Zbinden, K, Benz, J, Schlatter, D, Rudolph, M.G. | | Deposit date: | 2022-01-21 | | Release date: | 2022-10-12 | | Last modified: | 2025-08-06 | | Method: | X-RAY DIFFRACTION (2.19 Å) | | Cite: | A high quality, industrial data set for binding affinity prediction: performance comparison in different early drug discovery scenarios.
J.Comput.Aided Mol.Des., 36, 2022
|
|
5SEV
 
 | | CRYSTAL STRUCTURE OF HUMAN PHOSPHODIESTERASE 10 IN COMPLEX WITH c1(c(C)nc(c(n1)CCc2nc(cn2C)c3ccccc3)C)C, micromolar IC50=0.081773 | | Descriptor: | 2,3,5-trimethyl-6-[2-(1-methyl-4-phenyl-1H-imidazol-2-yl)ethyl]pyrazine, MAGNESIUM ION, ZINC ION, ... | | Authors: | Joseph, C, Groebke-Zbinden, K, Benz, J, Schlatter, D, Rudolph, M.G. | | Deposit date: | 2022-01-21 | | Release date: | 2022-10-12 | | Last modified: | 2025-08-06 | | Method: | X-RAY DIFFRACTION (2.31 Å) | | Cite: | A high quality, industrial data set for binding affinity prediction: performance comparison in different early drug discovery scenarios.
J.Comput.Aided Mol.Des., 36, 2022
|
|
5SF9
 
 | | CRYSTAL STRUCTURE OF HUMAN PHOSPHODIESTERASE 10 IN COMPLEX WITH c1ccc2c(c1)nc(c(n2)C)OCc3nc(cn3C)c4ccccc4, micromolar IC50=0.004192 | | Descriptor: | 2-methyl-3-[(1-methyl-4-phenyl-1H-imidazol-2-yl)methoxy]quinoxaline, MAGNESIUM ION, ZINC ION, ... | | Authors: | Joseph, C, Groebke-Zbinden, K, Benz, J, Schlatter, D, Rudolph, M.G. | | Deposit date: | 2022-01-21 | | Release date: | 2022-10-12 | | Last modified: | 2025-08-06 | | Method: | X-RAY DIFFRACTION (2.37 Å) | | Cite: | A high quality, industrial data set for binding affinity prediction: performance comparison in different early drug discovery scenarios.
J.Comput.Aided Mol.Des., 36, 2022
|
|
5SFA
 
 | | CRYSTAL STRUCTURE OF HUMAN PHOSPHODIESTERASE 10 IN COMPLEX WITH n1cc(nc(c1OCc2nc(cn2C)c3ccccc3)C(N[C@@H]4COCC4)=O)C5CC5, micromolar IC50=0.150444 | | Descriptor: | 6-cyclopropyl-3-[(1-methyl-4-phenyl-1H-imidazol-2-yl)methoxy]-N-[(3S)-oxolan-3-yl]pyrazine-2-carboxamide, MAGNESIUM ION, ZINC ION, ... | | Authors: | Joseph, C, Koerner, M, Benz, J, Schlatter, D, Rudolph, M.G. | | Deposit date: | 2022-01-21 | | Release date: | 2022-10-12 | | Last modified: | 2025-08-06 | | Method: | X-RAY DIFFRACTION (2.32 Å) | | Cite: | A high quality, industrial data set for binding affinity prediction: performance comparison in different early drug discovery scenarios.
J.Comput.Aided Mol.Des., 36, 2022
|
|
5SFC
 
 | | CRYSTAL STRUCTURE OF HUMAN PHOSPHODIESTERASE 10 IN COMPLEX WITH c1(c(cnn1C)C(Nc3nc2nc(cn2cc3)c4ccccc4)=O)C(N5CCC5)=O, micromolar IC50=2.191865 | | Descriptor: | 5-(azetidine-1-carbonyl)-1-methyl-N-[(4R)-2-phenylimidazo[1,2-a]pyrimidin-7-yl]-1H-pyrazole-4-carboxamide, MAGNESIUM ION, ZINC ION, ... | | Authors: | Joseph, C, Peters, J.U, Benz, J, Schlatter, D, Rudolph, M.G. | | Deposit date: | 2022-01-21 | | Release date: | 2022-10-12 | | Last modified: | 2025-08-06 | | Method: | X-RAY DIFFRACTION (2.18 Å) | | Cite: | A high quality, industrial data set for binding affinity prediction: performance comparison in different early drug discovery scenarios.
J.Comput.Aided Mol.Des., 36, 2022
|
|
5SFO
 
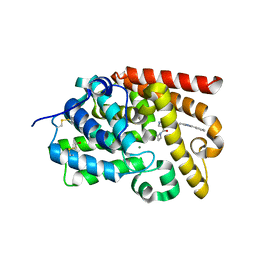 | | CRYSTAL STRUCTURE OF HUMAN PHOSPHODIESTERASE 10 IN COMPLEX WITH n2(cc(c1ccccc1)nc2CCNC(c4c(C(=O)N3CCC3)cnn4C)=O)C, micromolar IC50=0.004013 | | Descriptor: | 4-(azetidine-1-carbonyl)-1-methyl-N-[2-(1-methyl-4-phenyl-1H-imidazol-2-yl)ethyl]-1H-pyrazole-5-carboxamide, MAGNESIUM ION, ZINC ION, ... | | Authors: | Joseph, C, Koerner, M, Benz, J, Schlatter, D, Rudolph, M.G. | | Deposit date: | 2022-01-21 | | Release date: | 2022-10-12 | | Last modified: | 2025-08-06 | | Method: | X-RAY DIFFRACTION (1.96 Å) | | Cite: | A high quality, industrial data set for binding affinity prediction: performance comparison in different early drug discovery scenarios.
J.Comput.Aided Mol.Des., 36, 2022
|
|
5SJ4
 
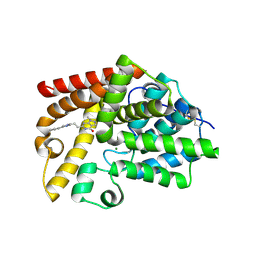 | | CRYSTAL STRUCTURE OF HUMAN PHOSPHODIESTERASE 10 IN COMPLEX WITH c1(ccccc1)c2cn(C)c(n2)CCC3=Nc4c(C(N3)=O)cnn4C, micromolar IC50=0.04979 | | Descriptor: | 1-methyl-6-[(E)-2-(1-methyl-4-phenyl-1H-imidazol-2-yl)ethen-1-yl]-1,5-dihydro-4H-pyrazolo[3,4-d]pyrimidin-4-one, MAGNESIUM ION, ZINC ION, ... | | Authors: | Joseph, C, Benz, J, Flohr, A, Groebke-Zbinden, K, Rudolph, M.G. | | Deposit date: | 2022-02-01 | | Release date: | 2022-10-12 | | Last modified: | 2025-08-06 | | Method: | X-RAY DIFFRACTION (2.77 Å) | | Cite: | Crystal Structure of a human phosphodiesterase 10 complex
To be published
|
|
5SJM
 
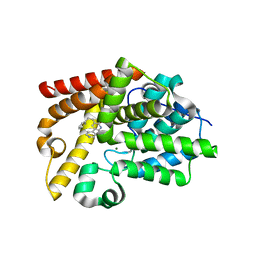 | | CRYSTAL STRUCTURE OF HUMAN PHOSPHODIESTERASE 10 IN COMPLEX WITH N1C(C(=CC(=C1)c2ccncc2)CCc3ccccc3)=O, micromolar IC50=1.981457 | | Descriptor: | 5-(2-phenylethyl)[3,4'-bipyridin]-6(1H)-one, MAGNESIUM ION, ZINC ION, ... | | Authors: | Joseph, C, Benz, J, Flohr, A, Bleicher, K, Rudolph, M.G. | | Deposit date: | 2022-02-01 | | Release date: | 2022-10-12 | | Last modified: | 2025-08-06 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystal Structure of a human phosphodiesterase 10 complex
To be published
|
|
5SJU
 
 | | CRYSTAL STRUCTURE OF HUMAN PHOSPHODIESTERASE 10 IN COMPLEX WITH c1c(nc(s1)NC(c2c(ncc(n2)C3CC3)Nc4cncnc4)=O)CN5CCCCC5, micromolar IC50=0.03208 | | Descriptor: | 6-cyclopropyl-N-{4-[(piperidin-1-yl)methyl]-1,3-thiazol-2-yl}-3-[(pyrimidin-5-yl)amino]pyrazine-2-carboxamide, MAGNESIUM ION, ZINC ION, ... | | Authors: | Joseph, C, Benz, J, Flohr, A, Koerner, M, Rudolph, M.G. | | Deposit date: | 2022-02-01 | | Release date: | 2022-10-12 | | Last modified: | 2025-08-06 | | Method: | X-RAY DIFFRACTION (1.97 Å) | | Cite: | Crystal Structure of a human phosphodiesterase 10 complex
To be published
|
|
5V93
 
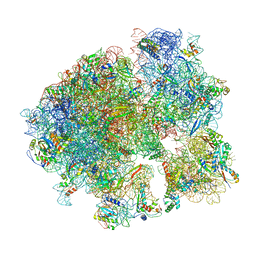 | | Cryo-EM structure of the 70S ribosome from Mycobacterium tuberculosis bound with Capreomycin | | Descriptor: | 16S rRNA, 23S rRNA, 30S ribosomal protein S10, ... | | Authors: | Yang, K, Chang, J.-Y, Cui, Z, Li, X, Meng, R, Duan, L, Thongchol, J, Jakana, J, Huwe, C, Sacchettini, J, Zhang, J. | | Deposit date: | 2017-03-22 | | Release date: | 2017-09-20 | | Last modified: | 2025-08-06 | | Method: | ELECTRON MICROSCOPY (4 Å) | | Cite: | Structural insights into species-specific features of the ribosome from the human pathogen Mycobacterium tuberculosis.
Nucleic Acids Res., 45, 2017
|
|
5W7T
 
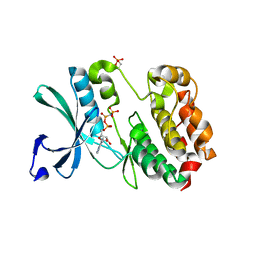 | | STRUCTURE OF PHOSPHORYLATED WNK1 | | Descriptor: | PHOSPHOAMINOPHOSPHONIC ACID-ADENYLATE ESTER, Serine/threonine-protein kinase WNK1 | | Authors: | JIOU, J, CHLEBOWICZ, J, AKELLA, R, GOLDSMITH, E.J. | | Deposit date: | 2017-06-20 | | Release date: | 2018-06-20 | | Last modified: | 2025-08-06 | | Method: | X-RAY DIFFRACTION (2.01 Å) | | Cite: | A Phosphorylated Intermediate in the Activation of WNK Kinases.
Biochemistry, 59, 2020
|
|
6YB7
 
 | | SARS-CoV-2 main protease with unliganded active site (2019-nCoV, coronavirus disease 2019, COVID-19). | | Descriptor: | 3C-like proteinase, DI(HYDROXYETHYL)ETHER, DIMETHYL SULFOXIDE | | Authors: | Owen, C.D, Lukacik, P, Strain-Damerell, C.M, Douangamath, A, Powell, A.J, Fearon, D, Brandao-Neto, J, Crawshaw, A.D, Aragao, D, Williams, M, Flaig, R, Hall, D.R, McAuley, K.E, Mazzorana, M, Stuart, D.I, von Delft, F, Walsh, M.A. | | Deposit date: | 2020-03-16 | | Release date: | 2020-03-25 | | Last modified: | 2025-08-06 | | Method: | X-RAY DIFFRACTION (1.25 Å) | | Cite: | Crystallographic and electrophilic fragment screening of the SARS-CoV-2 main protease.
Nat Commun, 11, 2020
|
|
7EZY
 
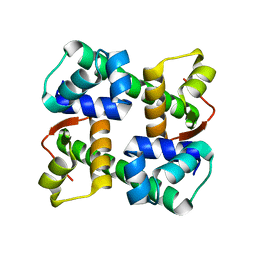 | | anti-CRISPR-associated Aca2 | | Descriptor: | anti-CRISPR-associated Aca2 | | Authors: | Lee, S.Y, Park, H.H. | | Deposit date: | 2021-06-02 | | Release date: | 2022-01-12 | | Last modified: | 2025-08-06 | | Method: | X-RAY DIFFRACTION (1.92 Å) | | Cite: | Molecular basis of transcriptional repression of anti-CRISPR by anti-CRISPR-associated 2.
Acta Crystallogr D Struct Biol, 78, 2022
|
|
7FW0
 
 | | Crystal Structure of human FABP4 in complex with 3-methyl-2-(2,4,5-trichlorophenyl)sulfanylbutanoic acid, i.e. SMILES c1(c(cc(c(c1)Cl)S[C@@H](C(=O)O)C(C)C)Cl)Cl with IC50=3.5 microM | | Descriptor: | (2R)-3-methyl-2-[(2,4,5-trichlorophenyl)sulfanyl]butanoic acid, (2S)-3-methyl-2-[(2,4,5-trichlorophenyl)sulfanyl]butanoic acid, FORMIC ACID, ... | | Authors: | Ehler, A, Benz, J, Obst, U, Richter, H, Rudolph, M.G. | | Deposit date: | 2023-04-27 | | Release date: | 2023-06-14 | | Last modified: | 2025-08-06 | | Method: | X-RAY DIFFRACTION (1.12 Å) | | Cite: | Crystal Structure of a human FABP4 complex
To be published
|
|
7FWC
 
 | | Crystal Structure of human FABP4 in complex with 5-(4-chlorophenyl)-2-(hydroxymethylene)cyclohexane-1,3-dione | | Descriptor: | 5-(4-chlorophenyl)-2-(hydroxymethylene)cyclohexane-1,3-dione, Fatty acid-binding protein, adipocyte, ... | | Authors: | Ehler, A, Benz, J, Obst, U, Rudolph, M.G. | | Deposit date: | 2023-04-27 | | Release date: | 2023-06-14 | | Last modified: | 2025-08-06 | | Method: | X-RAY DIFFRACTION (1.12 Å) | | Cite: | Crystal Structure of a human FABP4 complex
To be published
|
|
7FXC
 
 | | Crystal Structure of human FABP4 in complex with 6-[(3,4-dichlorophenyl)methyl]-5-methyl-1,1-dioxo-1,2,6-thiadiazin-3-one, i.e. SMILES N1(Cc2cc(c(cc2)Cl)Cl)S(=O)(=O)NC(=O)C=C1C with IC50=0.945 microM | | Descriptor: | 6-[(3,4-dichlorophenyl)methyl]-5-methyl-1lambda~6~,2,6-thiadiazine-1,1,3(2H,6H)-trione, Fatty acid-binding protein, adipocyte, ... | | Authors: | Ehler, A, Benz, J, Obst, U, Albrecht-Harry, A, Rudolph, M.G. | | Deposit date: | 2023-04-27 | | Release date: | 2023-06-14 | | Last modified: | 2025-08-06 | | Method: | X-RAY DIFFRACTION (1.12 Å) | | Cite: | Crystal Structure of a human FABP4 complex
To be published
|
|
7FXJ
 
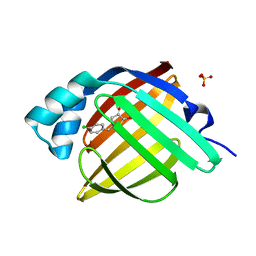 | | Crystal Structure of human FABP4 in complex with 1-[(4-chlorophenyl)methyl]-3,4-dihydroxy-2H-pyrrol-5-one | | Descriptor: | 1-[(4-chlorophenyl)methyl]-3,4-dihydroxy-1,5-dihydro-2H-pyrrol-2-one, Fatty acid-binding protein, adipocyte, ... | | Authors: | Ehler, A, Benz, J, Obst, U, Rudolph, M.G. | | Deposit date: | 2023-04-27 | | Release date: | 2023-06-14 | | Last modified: | 2025-08-06 | | Method: | X-RAY DIFFRACTION (1.03 Å) | | Cite: | Crystal Structure of a human FABP4 complex
To be published
|
|
7FY4
 
 | | Crystal Structure of human FABP4 binding site mutated to that of FABP5 in complex with 2-benzyl-6-tert-butyl-3-methyl-4-phenyl-5-(1H-tetrazol-5-yl)pyridine, i.e. SMILES N1C(=NN=N1)c1c(nc(c(c1c1ccccc1)C)Cc1ccccc1)C(C)(C)C with IC50=0.0608835 microM | | Descriptor: | (5M)-2-benzyl-6-tert-butyl-3-methyl-4-phenyl-5-(1H-tetrazol-5-yl)pyridine, Fatty acid-binding protein, adipocyte | | Authors: | Ehler, A, Benz, J, Obst, U, Buettelmann, B, Rudolph, M.G. | | Deposit date: | 2023-04-27 | | Release date: | 2023-06-14 | | Last modified: | 2025-08-06 | | Method: | X-RAY DIFFRACTION (1.51 Å) | | Cite: | Crystal Structure of a human FABP4 binding site mutated to that of FABP5 complex
To be published
|
|
