8WL4
 
 | |
8WM1
 
 | | DHS dehydratase | | Descriptor: | 3-dehydroshikimate dehydratase (DHS dehydratase), CALCIUM ION | | Authors: | Wang, M. | | Deposit date: | 2023-10-02 | | Release date: | 2024-10-02 | | Method: | X-RAY DIFFRACTION (2.74 Å) | | Cite: | DHS dehydratase
To Be Published
|
|
8WMB
 
 | |
8WME
 
 | |
8WSH
 
 | |
8WSJ
 
 | |
8WSK
 
 | |
8WWZ
 
 | |
8X6Q
 
 | | Crystal structure of OsHSL1 L204F/F298L/I335F complexed with 2-acetyl-cyclohexane-2,4-dione | | Descriptor: | 2-OXOGLUTARIC ACID, 2-ethanoyl-3-oxidanyl-cyclohex-2-en-1-one, COBALT (II) ION, ... | | Authors: | Lin, H.-Y, Dong, J, Yang, G.-F. | | Deposit date: | 2023-11-21 | | Release date: | 2024-10-02 | | Method: | X-RAY DIFFRACTION (2.39 Å) | | Cite: | An artificially evolved gene for herbicide-resistant rice breeding.
Proc.Natl.Acad.Sci.USA, 121, 2024
|
|
8X74
 
 | | Crystal structure of ZmHSL1A complexed with mesotrione | | Descriptor: | 2-OXOGLUTARIC ACID, 2-[(4-methylsulfonyl-2-nitro-phenyl)-oxidanyl-methylidene]cyclohexane-1,3-dione, 2-oxoglutarate (2OG) and Fe(II)-dependent oxygenase superfamily protein, ... | | Authors: | Lin, H.-Y, Dong, J, Yang, G.-F. | | Deposit date: | 2023-11-22 | | Release date: | 2024-10-02 | | Method: | X-RAY DIFFRACTION (1.791 Å) | | Cite: | An artificially evolved gene for herbicide-resistant rice breeding.
Proc.Natl.Acad.Sci.USA, 121, 2024
|
|
8X7C
 
 | | Crystal structure of ZmHSL1A | | Descriptor: | 2-OXOGLUTARIC ACID, 2-oxoglutarate (2OG) and Fe(II)-dependent oxygenase superfamily protein, COBALT (II) ION | | Authors: | Lin, H.-Y, Dong, J, Yang, G.-F. | | Deposit date: | 2023-11-23 | | Release date: | 2024-10-02 | | Method: | X-RAY DIFFRACTION (1.597 Å) | | Cite: | An artificially evolved gene for herbicide-resistant rice breeding.
Proc.Natl.Acad.Sci.USA, 121, 2024
|
|
8X7D
 
 | |
8XC3
 
 | | Crystal structure of ZmHSL1A-MBQ complex | | Descriptor: | 1,5-dimethyl-3-(2-methylphenyl)-6-(2-oxidanyl-6-oxidanylidene-cyclohexen-1-yl)carbonyl-quinazoline-2,4-dione, 2-OXOGLUTARIC ACID, 2-oxoglutarate (2OG) and Fe(II)-dependent oxygenase superfamily protein, ... | | Authors: | Yang, G.-F, Lin, H.-Y, Dong, J. | | Deposit date: | 2023-12-07 | | Release date: | 2024-10-02 | | Method: | X-RAY DIFFRACTION (1.79 Å) | | Cite: | An artificially evolved gene for herbicide-resistant rice breeding.
Proc.Natl.Acad.Sci.USA, 121, 2024
|
|
8XYR
 
 | | De novo designed protein GPX4-2 | | Descriptor: | De novo designed GPX4-2 | | Authors: | Liu, L.J, Guo, Z, Lai, L.H. | | Deposit date: | 2024-01-20 | | Release date: | 2024-10-02 | | Method: | X-RAY DIFFRACTION (1.87 Å) | | Cite: | All-Atom Protein Sequence Design Based on Geometric Deep Learning.
Angew.Chem.Int.Ed.Engl., 2024
|
|
8XYS
 
 | | De novo designed protein GPX4-1 | | Descriptor: | De novo designed GPX4-1 | | Authors: | Liu, J.L, Guo, Z, Lai, L.H. | | Deposit date: | 2024-01-20 | | Release date: | 2024-10-02 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | All-Atom Protein Sequence Design Based on Geometric Deep Learning.
Angew.Chem.Int.Ed.Engl., 2024
|
|
8XYT
 
 | | De novo designed protein GPX4-4 | | Descriptor: | De novo designed GPX4-4 | | Authors: | Liu, J.L, Guo, Z, Lai, L.H. | | Deposit date: | 2024-01-20 | | Release date: | 2024-10-02 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | All-Atom Protein Sequence Design Based on Geometric Deep Learning.
Angew.Chem.Int.Ed.Engl., 2024
|
|
8XYU
 
 | | De novo designed protein GPX4-3 | | Descriptor: | De novo designed GPX4-3 | | Authors: | Guo, Z, Liu, J.L, Lai, L.H. | | Deposit date: | 2024-01-20 | | Release date: | 2024-10-02 | | Method: | X-RAY DIFFRACTION (1.59 Å) | | Cite: | All-Atom Protein Sequence Design Based on Geometric Deep Learning.
Angew.Chem.Int.Ed.Engl., 2024
|
|
8XYV
 
 | | De novo designed protein 0705-5 | | Descriptor: | De novo designed protein 0705-5 | | Authors: | Liu, J.L, Guo, Z, Lai, L.H. | | Deposit date: | 2024-01-20 | | Release date: | 2024-10-02 | | Method: | X-RAY DIFFRACTION (1.81 Å) | | Cite: | All-Atom Protein Sequence Design Based on Geometric Deep Learning.
Angew.Chem.Int.Ed.Engl., 2024
|
|
8XYW
 
 | | De novo designed protein Trx-3 | | Descriptor: | De novo designed Trx-3 | | Authors: | Liu, J.L, Guo, Z, Lai, L.H. | | Deposit date: | 2024-01-20 | | Release date: | 2024-10-02 | | Method: | X-RAY DIFFRACTION (1.41 Å) | | Cite: | All-Atom Protein Sequence Design Based on Geometric Deep Learning.
Angew.Chem.Int.Ed.Engl., 2024
|
|
8YJN
 
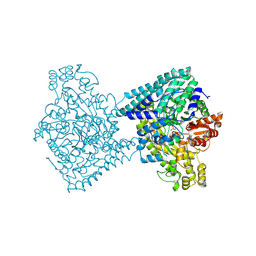 | | Structure of E. coli glycyl radical enzyme YbiW with bound glycerol | | Descriptor: | GLYCEROL, Probable dehydratase YbiW | | Authors: | Xue, B, Wei, Y, Robinson, R.C, Yew, W.S, Zhang, Y. | | Deposit date: | 2024-03-02 | | Release date: | 2024-10-02 | | Method: | X-RAY DIFFRACTION (2.42 Å) | | Cite: | A Widespread Radical-Mediated Glycolysis Pathway.
J.Am.Chem.Soc., 2024
|
|
8YJO
 
 | | Structure of E. coli glycyl radical enzyme PflD with bound malonate | | Descriptor: | MALONATE ION, Probable dehydratase PflD | | Authors: | Xue, B, Wei, Y, Robinson, R.C, Yew, W.S, Zhang, Y. | | Deposit date: | 2024-03-02 | | Release date: | 2024-10-02 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | A Widespread Radical-Mediated Glycolysis Pathway.
J.Am.Chem.Soc., 2024
|
|
8YSV
 
 | | Crystal structure of beta - glucosidase 6PG from Enterococcus faecalis | | Descriptor: | 1,2-ETHANEDIOL, 6-phospho-beta-glucosidase, ETHANOL, ... | | Authors: | Wang, W.Y, Li, Y.J, Liu, Z.Y, Han, X.D. | | Deposit date: | 2024-03-23 | | Release date: | 2024-10-02 | | Method: | X-RAY DIFFRACTION (1.79 Å) | | Cite: | Crystal structure of beta - glucosidase 6PG from Enterococcus faecalis
To Be Published
|
|
8Z2M
 
 | |
8Z2N
 
 | |
8Z2O
 
 | |
