1D6Z
 
 | | CRYSTAL STRUCTURE OF THE AEROBICALLY FREEZE TRAPPED RATE-DETERMINING CATALYTIC INTERMEDIATE OF E. COLI COPPER-CONTAINING AMINE OXIDASE. | | Descriptor: | 2-PHENYLETHYLAMINE, CALCIUM ION, COPPER (II) ION, ... | | Authors: | Wilmot, C.M, Hajdu, J, McPherson, M.J, Knowles, P.F, Phillips, S.E.V. | | Deposit date: | 1999-10-16 | | Release date: | 2000-02-02 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Visualization of dioxygen bound to copper during enzyme catalysis.
Science, 286, 1999
|
|
1D76
 
 | | CRYSTAL AND MOLECULAR STRUCTURE OF A DNA FRAGMENT CONTAINING A 2-AMINO ADENINE MODIFICATION: THE RELATIONSHIP BETWEEN CONFORMATION, PACKING, AND HYDRATION IN Z-DNA HEXAMERS | | Descriptor: | DNA (5'-D(*CP*GP*UP*(1AP)P*CP*G)-3') | | Authors: | Schneider, B, Ginell, S.L, Jones, R, Gaffney, B, Berman, H.M. | | Deposit date: | 1992-05-19 | | Release date: | 1992-09-15 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | Crystal and molecular structure of a DNA fragment containing a 2-aminoadenine modification: the relationship between conformation, packing, and hydration in Z-DNA hexamers.
Biochemistry, 31, 1992
|
|
1D78
 
 | | HIGH RESOLUTION REFINEMENT OF THE HEXAGONAL A-DNA OCTAMER D(GTGTACAC) AT 1.4 ANGSTROMS RESOLUTION | | Descriptor: | DNA (5'-D(*GP*TP*GP*TP*AP*CP*AP*C)-3') | | Authors: | Thota, N, Li, X.H, Bingman, C.A, Sundaralingam, M. | | Deposit date: | 1992-06-12 | | Release date: | 1993-04-15 | | Last modified: | 2023-03-22 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | High-resolution refinement of the hexagonal A-DNA octamer d(GTGTACAC) at 1.4 A.
Acta Crystallogr.,Sect.D, 49, 1993
|
|
1D79
 
 | | HIGH RESOLUTION REFINEMENT OF THE HEXAGONAL A-DNA OCTAMER D(GTGTACAC) AT 1.4 ANGSTROMS RESOLUTION | | Descriptor: | DNA (5'-D(*GP*TP*GP*TP*AP*CP*AP*C)-3') | | Authors: | Thota, N, Li, X.H, Bingman, C.A, Sundaralingam, M. | | Deposit date: | 1992-06-12 | | Release date: | 1993-04-15 | | Last modified: | 2023-03-22 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | High-resolution refinement of the hexagonal A-DNA octamer d(GTGTACAC) at 1.4 A.
Acta Crystallogr.,Sect.D, 49, 1993
|
|
1D7B
 
 | | CYTOCHROME DOMAIN OF CELLOBIOSE DEHYDROGENASE, PH 7.5 | | Descriptor: | 2-(2-{2-[2-(2-METHOXY-ETHOXY)-ETHOXY]-ETHOXY}-ETHOXY)-ETHANOL, CADMIUM ION, CELLOBIOSE DEHYDROGENASE, ... | | Authors: | Hallberg, B.M, Bergfors, T, Backbro, K, Divne, C. | | Deposit date: | 1999-10-16 | | Release date: | 2000-10-18 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | A new scaffold for binding haem in the cytochrome domain of the extracellular flavocytochrome cellobiose dehydrogenase.
Structure Fold.Des., 8, 2000
|
|
1D7C
 
 | | CYTOCHROME DOMAIN OF CELLOBIOSE DEHYDROGENASE, PH 4.6 | | Descriptor: | 2-(2-{2-[2-(2-METHOXY-ETHOXY)-ETHOXY]-ETHOXY}-ETHOXY)-ETHANOL, CADMIUM ION, CELLOBIOSE DEHYDROGENASE, ... | | Authors: | Hallberg, B.M, Bergfors, T, Backbro, K, Divne, C. | | Deposit date: | 1999-10-16 | | Release date: | 2000-10-18 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | A new scaffold for binding haem in the cytochrome domain of the extracellular flavocytochrome cellobiose dehydrogenase.
Structure Fold.Des., 8, 2000
|
|
1D7D
 
 | | CYTOCHROME DOMAIN OF CELLOBIOSE DEHYDROGENASE, HP3 FRAGMENT, PH 7.5 | | Descriptor: | 2-(2-{2-[2-(2-METHOXY-ETHOXY)-ETHOXY]-ETHOXY}-ETHOXY)-ETHANOL, CADMIUM ION, CELLOBIOSE DEHYDROGENASE, ... | | Authors: | Hallberg, B.M, Bergfors, T, Backbro, K, Divne, C. | | Deposit date: | 1999-10-16 | | Release date: | 2000-10-18 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | A new scaffold for binding haem in the cytochrome domain of the extracellular flavocytochrome cellobiose dehydrogenase.
Structure Fold.Des., 8, 2000
|
|
1D7F
 
 | | CRYSTAL STRUCTURE OF ASPARAGINE 233-REPLACED CYCLODEXTRIN GLUCANOTRANSFERASE FROM ALKALOPHILIC BACILLUS SP. 1011 DETERMINED AT 1.9 A RESOLUTION | | Descriptor: | CALCIUM ION, CYCLODEXTRIN GLUCANOTRANSFERASE | | Authors: | Ishii, N, Haga, K, Yamane, K, Harata, K. | | Deposit date: | 1999-10-18 | | Release date: | 2000-03-17 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal structure of asparagine 233-replaced cyclodextrin glucanotransferase from alkalophilic Bacillus sp. 1011 determined at 1.9 A resolution.
J.Mol.Recog., 13, 2000
|
|
1D7H
 
 | | FKBP COMPLEXED WITH DMSO | | Descriptor: | AMMONIUM ION, DIMETHYL SULFOXIDE, PROTEIN (FK506-BINDING PROTEIN), ... | | Authors: | Burkhard, P, Taylor, P, Walkinshaw, M.D. | | Deposit date: | 1999-10-18 | | Release date: | 1999-10-21 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | X-ray structures of small ligand-FKBP complexes provide an estimate for hydrophobic interaction energies.
J.Mol.Biol., 295, 2000
|
|
1D7I
 
 | | FKBP COMPLEXED WITH METHYL METHYLSULFINYLMETHYL SULFIDE (DSS) | | Descriptor: | AMMONIUM ION, METHYL METHYLSULFINYLMETHYL SULFIDE, PROTEIN (FK506-BINDING PROTEIN), ... | | Authors: | Burkhard, P, Taylor, P, Walkinshaw, M.D. | | Deposit date: | 1999-10-18 | | Release date: | 1999-10-21 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | X-ray structures of small ligand-FKBP complexes provide an estimate for hydrophobic interaction energies.
J.Mol.Biol., 295, 2000
|
|
1D7J
 
 | | FKBP COMPLEXED WITH 4-HYDROXY-2-BUTANONE | | Descriptor: | 4-HYDROXY-2-BUTANONE, AMMONIUM ION, PROTEIN (FK506-BINDING PROTEIN), ... | | Authors: | Burkhard, P, Taylor, P, Walkinshaw, M.D. | | Deposit date: | 1999-10-18 | | Release date: | 1999-10-21 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | X-ray structures of small ligand-FKBP complexes provide an estimate for hydrophobic interaction energies.
J.Mol.Biol., 295, 2000
|
|
1D7M
 
 | | COILED-COIL DIMERIZATION DOMAIN FROM CORTEXILLIN I | | Descriptor: | CORTEXILLIN I | | Authors: | Burkhard, P, Kammerer, R.A, Steinmetz, M.O, Bourenkov, G.P, Aebi, U. | | Deposit date: | 1999-10-19 | | Release date: | 2000-03-27 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | The coiled-coil trigger site of the rod domain of cortexillin I unveils a distinct network of interhelical and intrahelical salt bridges.
Structure Fold.Des., 8, 2000
|
|
1D7O
 
 | | CRYSTAL STRUCTURE OF BRASSICA NAPUS ENOYL ACYL CARRIER PROTEIN REDUCTASE COMPLEXED WITH NAD AND TRICLOSAN | | Descriptor: | ENOYL-[ACYL-CARRIER PROTEIN] REDUCTASE (NADH) PRECURSOR, NICOTINAMIDE-ADENINE-DINUCLEOTIDE, TRICLOSAN | | Authors: | Roujeinikova, A, Levy, C, Rowsell, S, Sedelnikova, S, Baker, P.J, Minshull, C.A, Mistry, A, Colls, J.G, Camble, R, Stuitje, A.R, Slabas, A.R, Rafferty, J.B, Pauptit, R.A, Viner, R, Rice, D.W. | | Deposit date: | 1999-10-19 | | Release date: | 1999-11-08 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystallographic analysis of triclosan bound to enoyl reductase.
J.Mol.Biol., 294, 1999
|
|
1D7W
 
 | |
1D7Z
 
 | |
1D80
 
 | |
1D89
 
 | |
1D8C
 
 | | MALATE SYNTHASE G COMPLEXED WITH MAGNESIUM AND GLYOXYLATE | | Descriptor: | GLYOXYLIC ACID, MAGNESIUM ION, MALATE SYNTHASE G, ... | | Authors: | Howard, B.R, Endrizzi, J.A, Remington, S.J. | | Deposit date: | 1999-10-22 | | Release date: | 1999-11-10 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal structure of Escherichia coli malate synthase G complexed with magnesium and glyoxylate at 2.0 A resolution: mechanistic implications.
Biochemistry, 39, 2000
|
|
1D8D
 
 | | CO-CRYSTAL STRUCTURE OF RAT PROTEIN FARNESYLTRANSFERASE COMPLEXED WITH A K-RAS4B PEPTIDE SUBSTRATE AND FPP ANALOG AT 2.0A RESOLUTION | | Descriptor: | ACETATE ION, K-RAS4B PEPTIDE SUBSTRATE, ZINC ION, ... | | Authors: | Long, S.B, Casey, P.J, Beese, L.S. | | Deposit date: | 1999-10-22 | | Release date: | 2000-02-09 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | The basis for K-Ras4B binding specificity to protein farnesyltransferase revealed by 2 A resolution ternary complex structures.
Structure Fold.Des., 8, 2000
|
|
1D8E
 
 | | Zinc-depleted FTase complexed with K-RAS4B peptide substrate and FPP analog. | | Descriptor: | ACETATE ION, FARNESYLTRANSFERASE (ALPHA SUBUNIT), FARNESYLTRANSFERASE (BETA SUBUNIT), ... | | Authors: | Long, S.B, Casey, P.J, Beese, L.S. | | Deposit date: | 1999-10-22 | | Release date: | 2000-03-20 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | The basis for K-Ras4B binding specificity to protein farnesyltransferase revealed by 2 A resolution ternary complex structures.
Structure Fold.Des., 8, 2000
|
|
1D8G
 
 | | ULTRAHIGH RESOLUTION CRYSTAL STRUCTURE OF B-DNA DECAMER D(CCAGTACTGG) | | Descriptor: | 5'-D(*CP*CP*AP*GP*TP*AP*CP*TP*GP*GP*)-3', CALCIUM ION | | Authors: | Kielkopf, C.L, Ding, S, Kuhn, P, Rees, D.C. | | Deposit date: | 1999-10-23 | | Release date: | 2000-02-21 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (0.74 Å) | | Cite: | Conformational flexibility of B-DNA at 0.74 A resolution: d(CCAGTACTGG)(2).
J.Mol.Biol., 296, 2000
|
|
1D8L
 
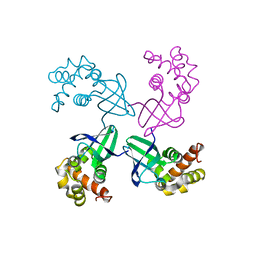 | | E. COLI HOLLIDAY JUNCTION BINDING PROTEIN RUVA NH2 REGION LACKING DOMAIN III | | Descriptor: | PROTEIN (HOLLIDAY JUNCTION DNA HELICASE RUVA) | | Authors: | Nishino, T, Iwasaki, H, Kataoka, M, Ariyoshi, M, Fujita, T, Shinagawa, H, Morikawa, K. | | Deposit date: | 1999-10-25 | | Release date: | 2000-05-03 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Modulation of RuvB function by the mobile domain III of the Holliday junction recognition protein RuvA.
J.Mol.Biol., 298, 2000
|
|
1D8S
 
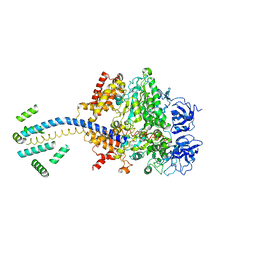 | | ESCHERICHIA COLI F1 ATPASE | | Descriptor: | F1 ATPASE (ALPHA SUBUNIT), F1 ATPASE (BETA SUBUNIT), F1 ATPASE (GAMMA SUBUNIT) | | Authors: | Hausrath, A.C, Gruber, G, Matthews, B.W, Capaldi, R.A. | | Deposit date: | 1999-10-25 | | Release date: | 1999-12-03 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (4.4 Å) | | Cite: | Structural features of the gamma subunit of the Escherichia coli F(1) ATPase revealed by a 4.4-A resolution map obtained by x-ray crystallography.
Proc.Natl.Acad.Sci.USA, 96, 1999
|
|
1D8U
 
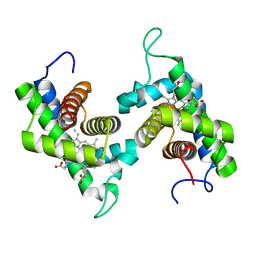 | | CRYSTAL STRUCTURE OF NON-SYMBIOTIC PLANT HEMOGLOBIN FROM RICE | | Descriptor: | NON-SYMBIOTIC HEMOGLOBIN, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Hargrove, M, Brucker, E.A, Stec, B, Olson, J.S, Phillips Jr, G.N. | | Deposit date: | 1999-10-26 | | Release date: | 2001-01-10 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Crystal structure of a nonsymbiotic plant hemoglobin.
Structure Fold.Des., 8, 2000
|
|
1D8W
 
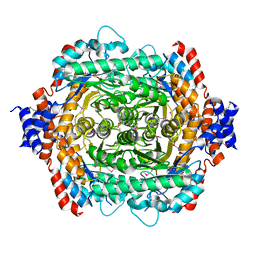 | | L-RHAMNOSE ISOMERASE | | Descriptor: | L-RHAMNOSE ISOMERASE, ZINC ION | | Authors: | Korndorfer, I.P, Matthews, B.W. | | Deposit date: | 1999-10-26 | | Release date: | 2000-09-27 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | The structure of rhamnose isomerase from Escherichia coli and its relation with xylose isomerase illustrates a change between inter and intra-subunit complementation during evolution.
J.Mol.Biol., 300, 2000
|
|
