1OXU
 
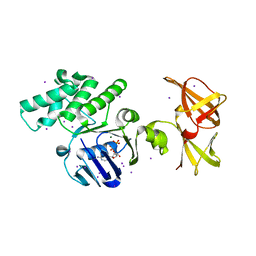 | | Crystal structure of GlcV, the ABC-ATPase of the glucose ABC transporter from Sulfolobus solfataricus | | Descriptor: | ABC transporter, ATP binding protein, ADENOSINE-5'-DIPHOSPHATE, ... | | Authors: | Verdon, G, Albers, S.V, Dijkstra, B.W, Driessen, A.J, Thunnissen, A.M. | | Deposit date: | 2003-04-03 | | Release date: | 2003-06-17 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Crystal structures of the ATPase subunit of the glucose ABC transporter from Sulfolobus solfataricus: nucleotide-free and nucleotide-bound conformations
J.Mol.Biol., 330, 2003
|
|
1OXV
 
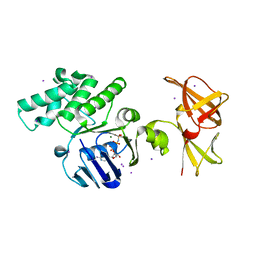 | | Crystal structure of GlcV, the ABC-ATPase of the glucose ABC transporter from Sulfolobus solfataricus | | Descriptor: | ABC transporter, ATP binding protein, IODIDE ION, ... | | Authors: | Verdon, G, Albers, S.V, Dijkstra, B.W, Driessen, A.J, Thunnissen, A.M. | | Deposit date: | 2003-04-03 | | Release date: | 2003-06-17 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Crystal structures of the ATPase subunit of the glucose ABC transporter from Sulfolobus solfataricus:
nucleotide-free and nucleotide-bound conformations
J.Mol.Biol., 330, 2003
|
|
1OXX
 
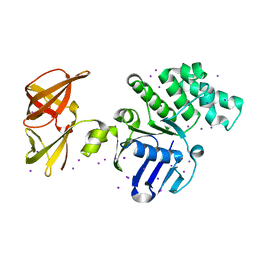 | | Crystal structure of GlcV, the ABC-ATPase of the glucose ABC transporter from Sulfolobus solfataricus | | Descriptor: | ABC transporter, ATP binding protein, IODIDE ION | | Authors: | Verdon, G, Albers, S.-V, van Oosterwijk, N, Dijkstra, B.W, Driessen, A.J.M, Thunnissen, A.M.W.H. | | Deposit date: | 2003-04-03 | | Release date: | 2003-09-30 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | Formation of the productive ATP-Mg2+-bound dimer of GlcV, an ABC-ATPase from Sulfolobus solfataricus
J.Mol.Biol., 334, 2003
|
|
1OXZ
 
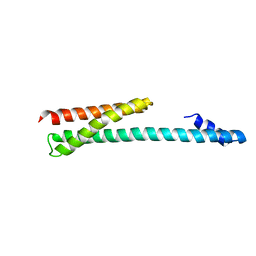 | | Crystal Structure of the Human GGA1 GAT domain | | Descriptor: | ADP-ribosylation factor binding protein GGA1 | | Authors: | Zhu, G, Zhai, P, He, X, Terzyan, S, Zhang, R, Joachimiak, A, Tang, J, Zhang, X.C. | | Deposit date: | 2003-04-03 | | Release date: | 2003-04-15 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Crystal Structure of Human GGA1 GAT Domain
Biochemistry, 42, 2003
|
|
1OY0
 
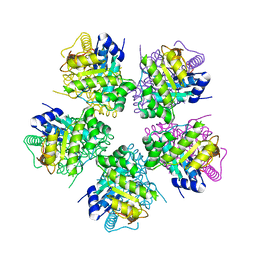 | | The crystal Structure of the First Enzyme of Pantothenate Biosynthetic Pathway, Ketopantoate Hydroxymethyltransferase from Mycobacterium Tuberculosis Shows a Decameric Assembly and Terminal Helix-Swapping | | Descriptor: | Ketopantoate hydroxymethyltransferase, MAGNESIUM ION | | Authors: | Chaudhuri, B.N, Sawaya, M.R, Kim, C.Y, Waldo, G.S, Park, M.S, Terwilliger, T.C, Yeates, T.O, TB Structural Genomics Consortium (TBSGC) | | Deposit date: | 2003-04-03 | | Release date: | 2003-07-15 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | The Crystal Structure of the First Enzyme in the Pantothenate Biosynthetic Pathway,
Ketopantoate Hydroxymethyltransferase, from M. tuberculosis
Structure, 11, 2003
|
|
1OY1
 
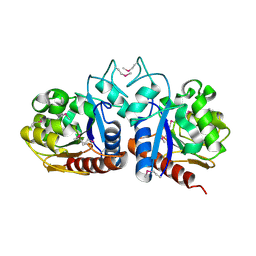 | | X-Ray Structure Of ElbB From E. Coli. Northeast Structural Genomics Research Consortium (Nesg) Target Er105 | | Descriptor: | PUTATIVE sigma cross-reacting protein 27A | | Authors: | Benach, J, Edstrom, W, Ma, L.C, Xiao, R, Acton, T.B, Rost, B, Montelione, G.T, Hunt, J.F, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2003-04-03 | | Release date: | 2003-04-22 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.95 Å) | | Cite: | X-Ray Structure Of ElbB From E. Coli. Northeast Structural Genomics Research Consortium (Nesg) Target Er105
To be Published
|
|
1OY3
 
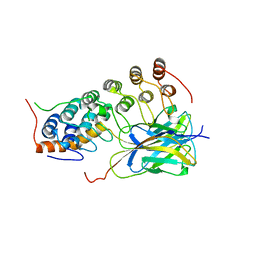 | | CRYSTAL STRUCTURE OF AN IKBBETA/NF-KB P65 HOMODIMER COMPLEX | | Descriptor: | Transcription factor p65, transcription factor inhibitor I-kappa-B-beta | | Authors: | Malek, S, Huang, D.B, Huxford, T, Ghosh, S, Ghosh, G. | | Deposit date: | 2003-04-03 | | Release date: | 2003-05-20 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | X-ray crystal structure of an IkappaBbeta x NF-kappaB p65 homodimer complex.
J.Biol.Chem., 278, 2003
|
|
1OY5
 
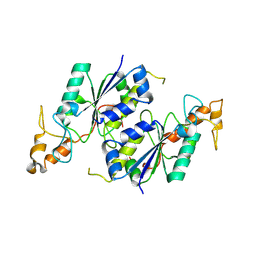 | | Crystal structure of tRNA (m1G37) methyltransferase from Aquifex aeolicus | | Descriptor: | tRNA (Guanine-N(1)-)-methyltransferase | | Authors: | Liu, J, Wang, W, Shin, D.H, Yokota, H, Kim, R, Kim, S.H, Berkeley Structural Genomics Center (BSGC) | | Deposit date: | 2003-04-03 | | Release date: | 2003-11-11 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Crystal structure of tRNA (m1G37) methyltransferase from Aquifex aeolicus at 2.6 A resolution: a novel methyltransferase fold.
Proteins, 53, 2003
|
|
1OY6
 
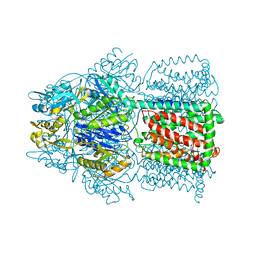 | | Structural Basis of the Multiple Binding Capacity of the AcrB Multidrug Efflux Pump | | Descriptor: | Acriflavine resistance protein B | | Authors: | Yu, E.W, McDermott, G, Zgurskaya, H.I, Nikaido, H, Koshland Jr, D.E. | | Deposit date: | 2003-04-03 | | Release date: | 2003-05-13 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (3.68 Å) | | Cite: | Structural basis of multiple drug-binding capacity of the AcrB multidrug efflux pump.
Science, 300, 2003
|
|
1OY7
 
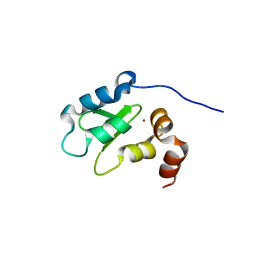 | | Structure and Function Analysis of Peptide Antagonists of Melanoma Inhibitor of Apoptosis (ML-IAP) | | Descriptor: | 3,6,9,12,15,18-HEXAOXAICOSANE-1,20-DIOL, AEVVAVKSE peptide, Baculoviral IAP repeat-containing protein 7, ... | | Authors: | Franklin, M.C, Kadkhodayan, S, Ackerly, H, Alexandru, D, Distefano, M.D, Elliott, L.O, Flygare, J.A, Vucic, D, Deshayes, K, Fairbrother, W.J. | | Deposit date: | 2003-04-03 | | Release date: | 2003-08-26 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Structure and Function Analysis of Peptide Antagonists of Melanoma Inhibitor of Apoptosis (ML-IAP)
Biochemistry, 42, 2003
|
|
1OY8
 
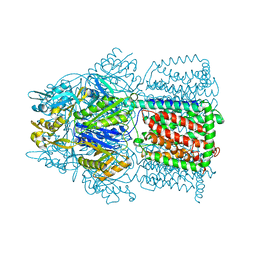 | | Structural Basis of Multiple Drug Binding Capacity of the AcrB Multidrug Efflux Pump | | Descriptor: | Acriflavine resistance protein B, RHODAMINE 6G | | Authors: | Yu, E.W, McDermott, G, Zgurskaya, H.I, Nikaido, H, Koshland Jr, D.E. | | Deposit date: | 2003-04-03 | | Release date: | 2003-05-13 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (3.63 Å) | | Cite: | Structural basis of multiple drug-binding capacity of the AcrB multidrug efflux pump.
Science, 300, 2003
|
|
1OY9
 
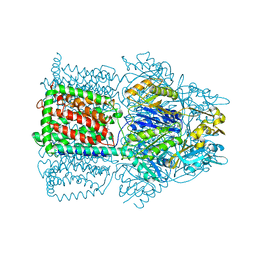 | | Structural Basis of Multiple Drug Binding Capacity of the AcrB Multidrug Efflux Pump | | Descriptor: | Acriflavine resistance protein B, ETHIDIUM | | Authors: | Yu, E.W, McDermott, G, Zgurskaya, H.I, Nikaido, H, Koshland Jr, D.E. | | Deposit date: | 2003-04-03 | | Release date: | 2003-05-13 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (3.8 Å) | | Cite: | Structural basis of multiple drug-binding capacity of the AcrB multidrug efflux pump.
Science, 300, 2003
|
|
1OYD
 
 | | Structural Basis of Multiple Binding Capacity of the AcrB multidrug Efflux Pump | | Descriptor: | Acriflavine resistance protein B, DEQUALINIUM | | Authors: | Yu, E.W, MeDermott, G, Zgurskaya, H.I, Nikaido, H, Koshland Jr, D.E. | | Deposit date: | 2003-04-03 | | Release date: | 2003-05-13 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (3.8 Å) | | Cite: | Structural basis of multiple drug-binding capacity of the AcrB multidrug efflux pump.
Science, 300, 2003
|
|
1OYE
 
 | | Structural Basis of Multiple Binding Capacity of the AcrB multidrug Efflux Pump | | Descriptor: | 1-CYCLOPROPYL-6-FLUORO-4-OXO-7-PIPERAZIN-1-YL-1,4-DIHYDROQUINOLINE-3-CARBOXYLIC ACID, Acriflavine resistance protein B | | Authors: | Yu, E.W, McDermott, G, Zgurskaya, H.I, Nikaido, H, Koshland Jr, D.E. | | Deposit date: | 2003-04-03 | | Release date: | 2003-05-13 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (3.48 Å) | | Cite: | Structural basis of multiple drug-binding capacity of the AcrB multidrug efflux pump.
Science, 300, 2003
|
|
1OYG
 
 | |
1OYH
 
 | | Crystal Structure of P13 Alanine Variant of Antithrombin | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Antithrombin-III, ... | | Authors: | Johnson, D.J.D, Huntington, J.A. | | Deposit date: | 2003-04-04 | | Release date: | 2004-04-13 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2.62 Å) | | Cite: | The influence of hinge region residue Glu-381 on antithrombin allostery and metastability
J.Biol.Chem., 279, 2004
|
|
1OYJ
 
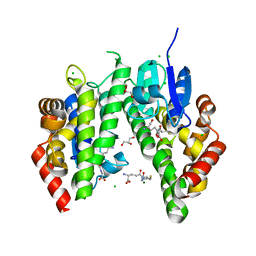 | | Crystal structure solution of Rice GST1 (OsGSTU1) in complex with glutathione. | | Descriptor: | CHLORIDE ION, GLUTATHIONE, GLYCEROL, ... | | Authors: | Dixon, D.P, McEwen, A.G, Lapthorn, A.J, Edwards, R. | | Deposit date: | 2003-04-04 | | Release date: | 2003-07-01 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Forced evolution of a herbicide detoxifying glutathione transferase.
J.Biol.Chem., 278, 2003
|
|
1OYN
 
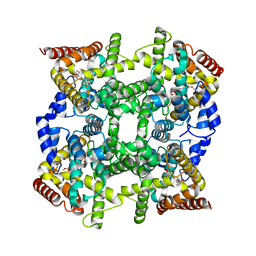 | | Crystal structure of PDE4D2 in complex with (R,S)-rolipram | | Descriptor: | ROLIPRAM, ZINC ION, cAMP-specific phosphodiesterase PDE4D2 | | Authors: | Huai, Q, Wang, H, Sun, Y, Kim, H.Y, Liu, Y, Ke, H. | | Deposit date: | 2003-04-05 | | Release date: | 2003-07-15 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Three-dimensional structures of PDE4D in complex with roliprams and implication on inhibitor selectivity
Structure, 11, 2003
|
|
1OYP
 
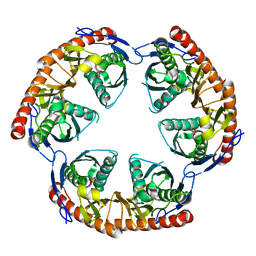 | | Crystal Structure of the phosphorolytic exoribonuclease RNase PH from Bacillus subtilis | | Descriptor: | Ribonuclease PH, SULFATE ION | | Authors: | Harlow, L.S, Kadziola, A, Jensen, K.F, Larsen, S. | | Deposit date: | 2003-04-07 | | Release date: | 2004-03-09 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.76 Å) | | Cite: | Crystal structure of the phosphorolytic exoribonuclease RNase PH from Bacillus subtilis and implications for its quaternary structure and tRNA binding.
Protein Sci., 13, 2004
|
|
1OYQ
 
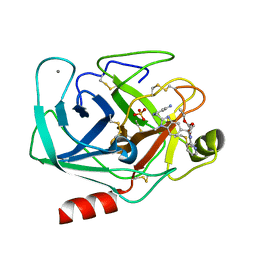 | | TRYPSIN INHIBITOR COMPLEX | | Descriptor: | CALCIUM ION, SULFATE ION, Trypsin, ... | | Authors: | Nar, H. | | Deposit date: | 2003-04-07 | | Release date: | 2003-04-29 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structural Basis for Inhibition Promiscuity of Dual Specific Thrombin and Factor Xa Blood Coagulation Inhibitors
Structure, 9, 2001
|
|
1OYR
 
 | | Crystal structure of the phosphorolytic exoribonuclease RNase PH from Bacillus subtilis | | Descriptor: | CADMIUM ION, Ribonuclease PH, SULFATE ION | | Authors: | Harlow, L.S, Kadziola, A, Jensen, K.F, Larsen, S. | | Deposit date: | 2003-04-07 | | Release date: | 2004-03-09 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Crystal structure of the phosphorolytic exoribonuclease RNase PH from Bacillus subtilis and implications for its quaternary structure and tRNA binding.
Protein Sci., 13, 2004
|
|
1OYS
 
 | |
1OYT
 
 | | COMPLEX OF RECOMBINANT HUMAN THROMBIN WITH A DESIGNED FLUORINATED INHIBITOR | | Descriptor: | (3ASR,4RS,8ASR,8BRS)-4-(2-(4-FLUOROBENZYL)-1,3-DIOXODEACAHYDROPYRROLO[3,4-A] PYRROLIZIN-4-YL)BENZAMIDINE, CALCIUM ION, Hirudin IIB, ... | | Authors: | Banner, D.W, Olsen, J.A. | | Deposit date: | 2003-04-07 | | Release date: | 2003-06-24 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.67 Å) | | Cite: | A Fluorine Scan of Thrombin Inhibitors to Map the Fluorophilicity/Fluorophobicity of an Enzyme Active Site: Evidence for CF...C=O Interactions.
Angew.Chem.Int.Ed.Engl., 42, 2003
|
|
1OYU
 
 | |
1OYV
 
 | | Crystal structure of tomato inhibitor-II in a ternary complex with subtilisin Carlsberg | | Descriptor: | CALCIUM ION, Subtilisin Carlsberg, Wound-induced proteinase inhibitor-II | | Authors: | Barrette-Ng, I.H, Ng, K.K, Cherney, M.M, Pearce, G, Ryan, C.A, James, M.N. | | Deposit date: | 2003-04-07 | | Release date: | 2003-07-15 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structural basis of inhibition revealed by a 1:2 complex of the two-headed tomato inhibitor-II and subtilisin Carlsberg
J.Biol.Chem., 278, 2003
|
|
