1B8G
 
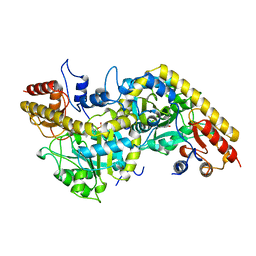 | | 1-AMINOCYCLOPROPANE-1-CARBOXYLATE SYNTHASE | | Descriptor: | PROTEIN (1-AMINOCYCLOPROPANE-1-CARBOXYLATE SYNTHASE), PYRIDOXAL-5'-PHOSPHATE | | Authors: | Capitani, G, Hohenester, E, Feng, L, Storici, P, Kirsch, J.F, Jansonius, J.N. | | Deposit date: | 1999-01-31 | | Release date: | 2000-01-26 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.37 Å) | | Cite: | Structure of 1-aminocyclopropane-1-carboxylate synthase, a key enzyme in the biosynthesis of the plant hormone ethylene.
J.Mol.Biol., 294, 1999
|
|
1B8H
 
 | | SLIDING CLAMP, DNA POLYMERASE | | Descriptor: | DNA POLYMERASE PROCESSIVITY COMPONENT, DNA POLYMERASE fragment | | Authors: | Shamoo, Y, Steitz, T.A. | | Deposit date: | 1999-02-01 | | Release date: | 1999-02-09 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Building a replisome from interacting pieces: sliding clamp complexed to a peptide from DNA polymerase and a polymerase editing complex.
Cell(Cambridge,Mass.), 99, 1999
|
|
1B8L
 
 | | Calcium-bound D51A/E101D/F102W Triple Mutant of Beta Carp Parvalbumin | | Descriptor: | CALCIUM ION, CARBONATE ION, PROTEIN (PARVALBUMIN) | | Authors: | Cates, M.S, Berry, M.B, Ho, E, Li, Q, Potter, J.D, Phillips Jr, G.N. | | Deposit date: | 1999-02-01 | | Release date: | 1999-10-05 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Metal-ion affinity and specificity in EF-hand proteins: coordination geometry and domain plasticity in parvalbumin.
Structure Fold.Des., 7, 1999
|
|
1B8R
 
 | | PARVALBUMIN | | Descriptor: | CALCIUM ION, PROTEIN (PARVALBUMIN) | | Authors: | Cates, M.S, Berry, M.B, Ho, E.L, Li, Q, Potter, J.D, Phillips Jr, G.N. | | Deposit date: | 1999-02-02 | | Release date: | 1999-02-09 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Metal-ion affinity and specificity in EF-hand proteins: coordination geometry and domain plasticity in parvalbumin.
Structure Fold.Des., 7, 1999
|
|
1B8Z
 
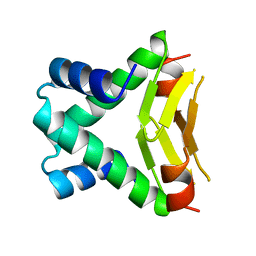 | | HU FROM THERMOTOGA MARITIMA | | Descriptor: | PROTEIN (HISTONELIKE PROTEIN HU) | | Authors: | Christodoulou, E, Rypniewski, W.R, Vorgias, C.E. | | Deposit date: | 1999-02-03 | | Release date: | 2000-02-03 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Cloning, overproduction, purification and crystallization of the DNA binding protein HU from the hyperthermophilic eubacterium Thermotoga maritima.
Acta Crystallogr.,Sect.D, 54, 1998
|
|
1B93
 
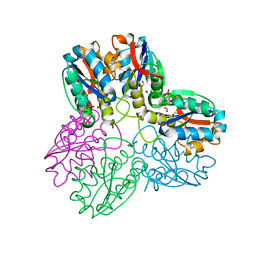 | | METHYLGLYOXAL SYNTHASE FROM ESCHERICHIA COLI | | Descriptor: | FORMIC ACID, PHOSPHATE ION, PROTEIN (METHYLGLYOXAL SYNTHASE) | | Authors: | Saadat, D, Harrison, D.H.T. | | Deposit date: | 1999-02-23 | | Release date: | 1999-03-16 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | The crystal structure of methylglyoxal synthase from Escherichia coli.
Structure Fold.Des., 7, 1999
|
|
1B94
 
 | | RESTRICTION ENDONUCLEASE ECORV WITH CALCIUM | | Descriptor: | CALCIUM ION, DNA (5'-D(*AP*AP*AP*GP*AP*TP*AP*TP*CP*TP*T)-3'), RESTRICTION ENDONUCLEASE ECORV | | Authors: | Thomas, M.P, Halford, S.E, Brady, R.L. | | Deposit date: | 1999-02-19 | | Release date: | 1999-02-26 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structural analysis of a mutational hot-spot in the EcoRV restriction endonuclease: a catalytic role for a main chain carbonyl group.
Nucleic Acids Res., 27, 1999
|
|
1B95
 
 | |
1B96
 
 | |
1B97
 
 | |
1B9A
 
 | | PARVALBUMIN (MUTATION;D51A, F102W) | | Descriptor: | CALCIUM ION, PROTEIN (PARVALBUMIN) | | Authors: | Cates, M.S, Berry, M.B, Ho, E.L, Li, Q, Potter, J.D, Phillips Jr, G.N. | | Deposit date: | 1999-02-10 | | Release date: | 1999-02-15 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Metal-ion affinity and specificity in EF-hand proteins: coordination geometry and domain plasticity in parvalbumin.
Structure Fold.Des., 7, 1999
|
|
1B9B
 
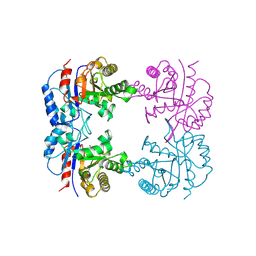 | | TRIOSEPHOSPHATE ISOMERASE OF THERMOTOGA MARITIMA | | Descriptor: | PROTEIN (TRIOSEPHOSPHATE ISOMERASE), SULFATE ION | | Authors: | Maes, D, Wierenga, R.K. | | Deposit date: | 1999-02-09 | | Release date: | 2000-01-01 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (2.85 Å) | | Cite: | The crystal structure of triosephosphate isomerase (TIM) from Thermotoga maritima: a comparative thermostability structural analysis of ten different TIM structures.
Proteins, 37, 1999
|
|
1B9C
 
 | |
1B9E
 
 | | HUMAN INSULIN MUTANT SERB9GLU | | Descriptor: | PROTEIN (INSULIN) | | Authors: | Wang, D.C, Zeng, Z.H, Yao, Z.P, Li, H.M. | | Deposit date: | 1998-11-12 | | Release date: | 1999-11-17 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structure of an insulin dimer in an orthorhombic crystal: the structure analysis of a human insulin mutant (B9 Ser-->Glu).
Acta Crystallogr.,Sect.D, 55, 1999
|
|
1B9H
 
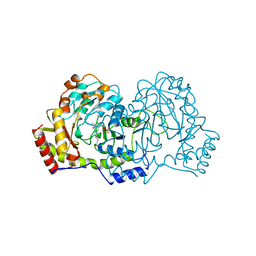 | | CRYSTAL STRUCTURE OF 3-AMINO-5-HYDROXYBENZOIC ACID (AHBA) SYNTHASE | | Descriptor: | PROTEIN (3-AMINO-5-HYDROXYBENZOIC ACID SYNTHASE), PYRIDOXAL-5'-PHOSPHATE | | Authors: | Eads, J.C, Beeby, M, Scapin, G, Yu, T.-W, Floss, H.G. | | Deposit date: | 1999-02-11 | | Release date: | 1999-08-13 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal structure of 3-amino-5-hydroxybenzoic acid (AHBA) synthase.
Biochemistry, 38, 1999
|
|
1B9J
 
 | |
1B9K
 
 | |
1B9M
 
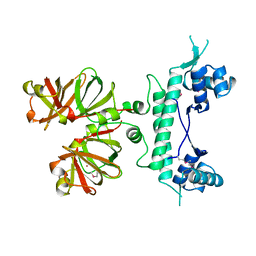 | | REGULATOR FROM ESCHERICHIA COLI | | Descriptor: | NICKEL (II) ION, PROTEIN (MODE) | | Authors: | Hall, D.R, Gourley, D.G, Hunter, W.N. | | Deposit date: | 1999-02-12 | | Release date: | 2000-03-15 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | The high-resolution crystal structure of the molybdate-dependent transcriptional regulator (ModE) from Escherichia coli: a novel combination of domain folds.
EMBO J., 18, 1999
|
|
1B9N
 
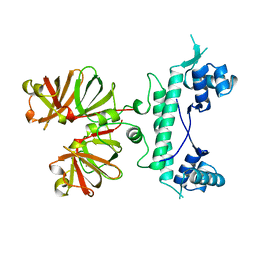 | | REGULATOR FROM ESCHERICHIA COLI | | Descriptor: | NICKEL (II) ION, PROTEIN (MODE) | | Authors: | Hall, D.R, Gourley, D.G, Hunter, W.N. | | Deposit date: | 1999-02-12 | | Release date: | 2000-03-15 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.09 Å) | | Cite: | The high-resolution crystal structure of the molybdate-dependent transcriptional regulator (ModE) from Escherichia coli: a novel combination of domain folds.
EMBO J., 18, 1999
|
|
1B9O
 
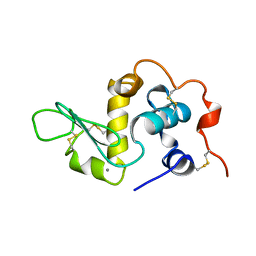 | | HUMAN ALPHA-LACTALBUMIN, LOW TEMPERATURE FORM | | Descriptor: | CALCIUM ION, PROTEIN (ALPHA-LACTALBUMIN) | | Authors: | Harata, K, Abe, Y, Muraki, M. | | Deposit date: | 1999-02-14 | | Release date: | 1999-03-31 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.15 Å) | | Cite: | Crystallographic evaluation of internal motion of human alpha-lactalbumin refined by full-matrix least-squares method.
J.Mol.Biol., 287, 1999
|
|
1B9S
 
 | | NOVEL AROMATIC INHIBITORS OF INFLUENZA VIRUS NEURAMINIDASE MAKE SELECTIVE INTERACTIONS WITH CONSERVED RESIDUES AND WATER MOLECULES IN THE ACTIVE SITE | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 4-(N-ACETYLAMINO)-3-[N-(2-ETHYLBUTANOYLAMINO)]BENZOIC ACID, CALCIUM ION, ... | | Authors: | Finley, J.B, Atigadda, V.R, Duarte, F, Zhao, J.J, Brouillette, W.J, Air, G.M, Luo, M. | | Deposit date: | 1999-02-15 | | Release date: | 1999-02-26 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Novel aromatic inhibitors of influenza virus neuraminidase make selective interactions with conserved residues and water molecules in the active site.
J.Mol.Biol., 293, 1999
|
|
1B9T
 
 | | NOVEL AROMATIC INHIBITORS OF INFLUENZA VIRUS NEURAMINIDASE MAKE SELECTIVE INTERACTIONS WITH CONSERVED RESIDUES AND WATER MOLECULES IN THE ACTIVE SITE | | Descriptor: | 1-(4-CARBOXY-2-GUANIDINOPENTYL)-5,5'-DI(HYDROXYMETHYL)PYRROLIDIN-2-ONE, 2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, ... | | Authors: | Finley, J.B, Atigadda, V.R, Duarte, F, Zhao, J.J, Brouillette, W.J, Air, G.M, Luo, M. | | Deposit date: | 1999-02-15 | | Release date: | 1999-02-27 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Novel aromatic inhibitors of influenza virus neuraminidase make selective interactions with conserved residues and water molecules in the active site.
J.Mol.Biol., 293, 1999
|
|
1B9V
 
 | | NOVEL AROMATIC INHIBITORS OF INFLUENZA VIRUS NEURAMINIDASE MAKE SELECTIVE INTERACTIONS WITH CONSERVED RESIDUES AND WATER MOLECULES IN TEH ACTIVE SITE | | Descriptor: | 1-[4-CARBOXY-2-(3-PENTYLAMINO)PHENYL]-5,5'-DI(HYDROXYMETHYL)PYRROLIDIN-2-ONE, 2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, ... | | Authors: | Finley, J.B, Atigadda, V.R, Duarte, F, Zahao, J.J, Brouillette, W.J, Air, G.M, Luo, M. | | Deposit date: | 1999-02-15 | | Release date: | 1999-02-27 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Novel aromatic inhibitors of influenza virus neuraminidase make selective interactions with conserved residues and water molecules in the active site.
J.Mol.Biol., 293, 1999
|
|
1B9W
 
 | | C-TERMINAL MEROZOITE SURFACE PROTEIN 1 FROM PLASMODIUM CYNOMOLGI | | Descriptor: | PROTEIN (MEROZOITE SURFACE PROTEIN 1) | | Authors: | Bentley, G.A, Chitarra, V, Holm, I, Longacre, S. | | Deposit date: | 1999-02-15 | | Release date: | 1999-05-24 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | The crystal structure of C-terminal merozoite surface protein 1 at 1.8 A resolution, a highly protective malaria vaccine candidate.
Mol.Cell, 3, 1999
|
|
1B9X
 
 | |
