1PU7
 
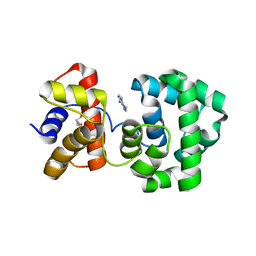 | | Crystal structure of H.pylori 3-methyladenine DNA glycosylase (MagIII) bound to 3,9-dimethyladenine | | Descriptor: | 3-METHYLADENINE DNA GLYCOSYLASE, 6-AMINO-3,9-DIMETHYL-9H-PURIN-3-IUM, BETA-MERCAPTOETHANOL | | Authors: | Eichman, B.F, O'Rourke, E.J, Radicella, J.P, Ellenberger, T. | | Deposit date: | 2003-06-24 | | Release date: | 2003-10-07 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.93 Å) | | Cite: | Crystal structures of 3-methyladenine DNA glycosylase MagIII and the recognition of alkylated bases
Embo J., 22, 2003
|
|
1PU8
 
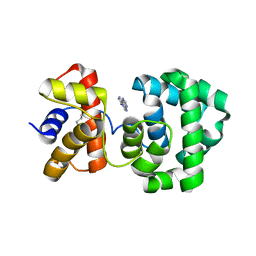 | | Crystal structure of H.pylori 3-methyladenine DNA glycosylase (MagIII) bound to 1,N6-ethenoadenine | | Descriptor: | 3-METHYLADENINE DNA GLYCOSYLASE, 3H-IMIDAZO[2,1-I]PURINE, BETA-MERCAPTOETHANOL | | Authors: | Eichman, B.F, O'Rourke, E.J, Radicella, J.P, Ellenberger, T. | | Deposit date: | 2003-06-24 | | Release date: | 2003-10-07 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.13 Å) | | Cite: | Crystal structures of 3-methyladenine DNA glycosylase MagIII and the recognition of alkylated bases
Embo J., 22, 2003
|
|
1PUB
 
 | |
1PUC
 
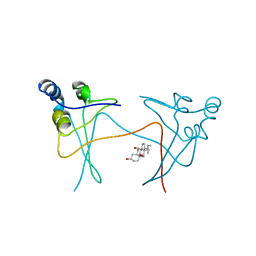 | | P13SUC1 IN A STRAND-EXCHANGED DIMER | | Descriptor: | 3-[(3-CHOLAMIDOPROPYL)DIMETHYLAMMONIO]-1-PROPANESULFONATE, P13SUC1 | | Authors: | Khazanovich, N, Bateman, K.S, Chernaia, M, Michalak, M, James, M.N.G. | | Deposit date: | 1995-12-08 | | Release date: | 1996-04-03 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Crystal structure of the yeast cell-cycle control protein, p13suc1, in a strand-exchanged dimer.
Structure, 4, 1996
|
|
1PUF
 
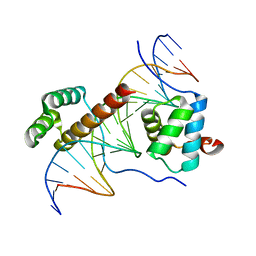 | | Crystal Structure of HoxA9 and Pbx1 homeodomains bound to DNA | | Descriptor: | 5'-D(*AP*CP*TP*CP*TP*AP*TP*GP*AP*TP*TP*TP*AP*CP*GP*AP*CP*GP*CP*T)-3', 5'-D(*TP*AP*GP*CP*GP*TP*CP*GP*TP*AP*AP*AP*TP*CP*AP*TP*AP*GP*AP*G)-3', Homeobox protein Hox-A9, ... | | Authors: | Laronde-Leblanc, N.A, Wolberger, C. | | Deposit date: | 2003-06-24 | | Release date: | 2003-09-02 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | STRUCTURE OF HOXA9 AND PBX1 BOUND TO DNA: HOX HEXAPEPTIDE AND DNA RECOGNITION ANTERIOR TO POSTERIOR
Genes Dev., 17, 2003
|
|
1PUG
 
 | | Structure of E. coli Ybab | | Descriptor: | Hypothetical UPF0133 protein ybaB | | Authors: | Kniewel, R, Buglino, J, Chadna, T, Lima, C.D, Burley, S.K, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2003-06-24 | | Release date: | 2003-07-08 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structure of E. coli Ybab
To be Published
|
|
1PUI
 
 | |
1PUJ
 
 | | Structure of B. subtilis YlqF GTPase | | Descriptor: | MAGNESIUM ION, PHOSPHOAMINOPHOSPHONIC ACID-GUANYLATE ESTER, conserved hypothetical protein ylqF | | Authors: | Kniewel, R, Buglino, J, Lima, C.D, Burley, S.K, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2003-06-24 | | Release date: | 2003-07-08 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structure of the YlqF GTPase from B. subtilis
To be Published
|
|
1PUM
 
 | | Mistletoe lectin I in complex with galactose | | Descriptor: | 2-acetamido-2-deoxy-alpha-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Krauspenhaar, R, Voelter, W, Stoeva, S, Mikhailov, A, Konareva, N, Betzel, C. | | Deposit date: | 2003-06-25 | | Release date: | 2004-06-25 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Mistletoe lectin I in complex with galactose and lactose reveals distinct sugar-binding properties
Acta Crystallogr.,Sect.F, 61, 2005
|
|
1PUO
 
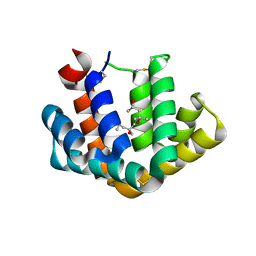 | | Crystal structure of Fel d 1- the major cat allergen | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, Major allergen I polypeptide, fused chain 2, ... | | Authors: | Kaiser, L, Gronlund, H, Sandalova, T, Ljunggren, H.G, van Hage-Hamsten, M, Achour, A, Schneider, G. | | Deposit date: | 2003-06-25 | | Release date: | 2003-10-14 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | The crystal structure of the major cat allergen Fel d 1, a member of the secretoglobin family.
J.Biol.Chem., 278, 2003
|
|
1PUU
 
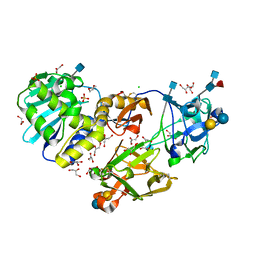 | | Mistletoe lectin I in complex with lactose | | Descriptor: | 1,4-DIETHYLENE DIOXIDE, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Krauspenhaar, R, Voelter, W, Stoeva, S, Mikhailov, A, Konareva, N, Betzel, C. | | Deposit date: | 2003-06-25 | | Release date: | 2004-06-25 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Mistletoe lectin I in complex with galactose and lactose reveals distinct sugar-binding properties
Acta Crystallogr.,Sect.F, 61, 2005
|
|
1PUY
 
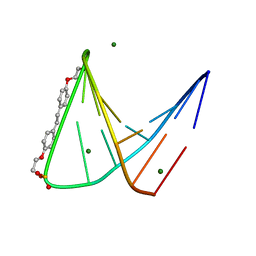 | | 1.5 A resolution structure of a synthetic DNA hairpin with a stilbenediether linker | | Descriptor: | 5'-D(*GP*TP*TP*TP*TP*GP*(S02)P*CP*AP*AP*AP*AP*C)-3', MAGNESIUM ION | | Authors: | Egli, M, Tereshko, V, Murshudov, G, Sanishvili, R, Liu, X, Lewis, F.D. | | Deposit date: | 2003-06-25 | | Release date: | 2003-10-14 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Face-to-face and edge-to-face pi-pi interactions in a synthetic DNA hairpin with a stilbenediether linker
J.Am.Chem.Soc., 125, 2003
|
|
1PV1
 
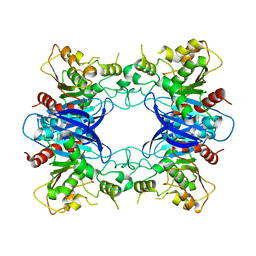 | | Crystal Structure Analysis of Yeast Hypothetical Protein: YJG8_YEAST | | Descriptor: | Hypothetical 33.9 kDa esterase in SMC3-MRPL8 intergenic region | | Authors: | Millard, C, Kumaran, D, Eswaramoorthy, S, Swaminathan, S, Burley, S.K, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2003-06-26 | | Release date: | 2004-11-30 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structural characterization and reversal of the natural organophosphate resistance of a D-type esterase, Saccharomyces cerevisiae S-formylglutathione hydrolase.
Biochemistry, 47, 2008
|
|
1PV4
 
 | |
1PV5
 
 | |
1PV8
 
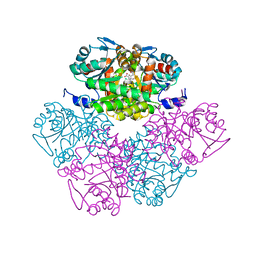 | | Crystal structure of a low activity F12L mutant of human porphobilinogen synthase | | Descriptor: | 3-(2-AMINOETHYL)-4-(AMINOMETHYL)HEPTANEDIOIC ACID, Delta-aminolevulinic acid dehydratase, ZINC ION | | Authors: | Breinig, S, Kervinen, J, Stith, L, Wasson, A.S, Fairman, R, Wlodawer, A, Zdanov, A, Jaffe, E.K. | | Deposit date: | 2003-06-26 | | Release date: | 2003-09-09 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Control of tetrapyrrole biosynthesis by alternate quaternary forms of porphobilinogen synthase.
Nat.Struct.Biol., 10, 2003
|
|
1PV9
 
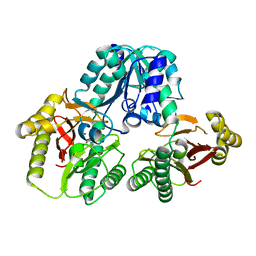 | | Prolidase from Pyrococcus furiosus | | Descriptor: | Xaa-Pro dipeptidase, ZINC ION | | Authors: | Maher, M.J, Ghosh, M, Grunden, A.M, Menon, A.L, Adams, M.W, Freeman, H.C, Guss, J.M. | | Deposit date: | 2003-06-27 | | Release date: | 2004-03-23 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structure of the Prolidase from Pyrococcus furiosus.
Biochemistry, 43, 2004
|
|
1PVA
 
 | | COMPARISON BETWEEN THE CRYSTAL AND THE SOLUTION STRUCTURES OF THE EF HAND PARVALBUMIN (ALPHA COMPONENT FROM PIKE MUSCLE) | | Descriptor: | CALCIUM ION, PARVALBUMIN | | Authors: | Declercq, J.P, Tinant, B, Roquet, F, Rambaud, J, Parello, J. | | Deposit date: | 1995-01-16 | | Release date: | 1995-03-31 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Comparison between the Crystal and the Solution Structures of the EF Hand Parvalbumin (Alpha Component from Pike Muscle)
To be Published
|
|
1PVB
 
 | | X-RAY STRUCTURE OF A NEW CRYSTAL FORM OF PIKE 4.10 PARVALBUMIN | | Descriptor: | AMMONIUM ION, CALCIUM ION, PARVALBUMIN | | Authors: | Declercq, J.P, Tinant, B, Parello, J. | | Deposit date: | 1995-01-05 | | Release date: | 1995-02-27 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | X-ray structure of a new crystal form of pike 4.10 beta parvalbumin.
Acta Crystallogr.,Sect.D, 52, 1996
|
|
1PVD
 
 | |
1PVF
 
 | | E.coli IPP isomerase in complex with diphosphate | | Descriptor: | DIPHOSPHATE, Isopentenyl-diphosphate delta-isomerase, MAGNESIUM ION, ... | | Authors: | Wouters, J, Durisotti, V, Oudjama, Y, Stalon, V, Droogmans, L. | | Deposit date: | 2003-06-27 | | Release date: | 2004-07-06 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.78 Å) | | Cite: | E.coli Isopentenyl Diphosphate: Dimethylallyl Diphosphate isomerase in complex with diphosphate
To be Published
|
|
1PVG
 
 | |
1PVH
 
 | | Crystal structure of leukemia inhibitory factor in complex with gp130 | | Descriptor: | IODIDE ION, Interleukin-6 receptor beta chain, Leukemia inhibitory factor | | Authors: | Boulanger, M.J, Bankovich, A.J, Kortemme, T, Baker, D, Garcia, K.C. | | Deposit date: | 2003-06-27 | | Release date: | 2003-10-14 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Convergent mechanisms for recognition of divergent cytokines by the shared signaling receptor gp130.
Mol.Cell, 12, 2003
|
|
1PVJ
 
 | | Crystal structure of the Streptococcal pyrogenic exotoxin B (SpeB)- inhibitor complex | | Descriptor: | (3R)-3-{[(BENZYLOXY)CARBONYL]AMINO}-2-OXO-4-PHENYLBUTANE-1-DIAZONIUM, pyrogenic exotoxin B | | Authors: | Ziomek, E, Sivaraman, J, Doran, J, Menard, R, Cygler, M. | | Deposit date: | 2003-06-27 | | Release date: | 2004-09-28 | | Last modified: | 2017-10-11 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Inhibition of autoprocessing of the streptococcal pyrogenic exotoxin B (speB). Crystal structure of the proenzyme-inhibitor complex
To be published
|
|
1PVL
 
 | | STRUCTURE OF THE PANTON-VALENTINE LEUCOCIDIN F COMPONENT FROM STAPHYLOCOCCUS AUREUS | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, LEUCOCIDIN | | Authors: | Pedelacq, J.D, Mourey, L, Maveyraud, L, Prevost, G, Samama, J.P. | | Deposit date: | 1999-01-12 | | Release date: | 1999-06-15 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | The structure of a Staphylococcus aureus leucocidin component (LukF-PV) reveals the fold of the water-soluble species of a family of transmembrane pore-forming toxins.
Structure Fold.Des., 7, 1999
|
|
