1OSC
 
 | | Crystal structure of rat CUTA1 at 2.15 A resolution | | Descriptor: | similar to divalent cation tolerant protein CUTA | | Authors: | Arnesano, F, Banci, L, Benvenuti, M, Bertini, I, Calderone, V, Mangani, S, Viezzoli, M.S, Structural Proteomics in Europe (SPINE) | | Deposit date: | 2003-03-19 | | Release date: | 2003-11-25 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | The Evolutionarily Conserved Trimeric Structure of CutA1 Proteins
Suggests a Role in Signal Transduction
J.Biol.Chem., 278, 2003
|
|
1OSD
 
 | | crystal structure of Oxidized MerP from Ralstonia metallidurans CH34 | | Descriptor: | hypothetical protein MerP | | Authors: | Serre, L, Rossy, E, Pebay-Peyroula, E, Cohen-Addad, C, Coves, J. | | Deposit date: | 2003-03-19 | | Release date: | 2004-05-18 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal Structure of the Oxidized Form of the Periplasmic Mercury-binding Protein MerP from Ralstonia metallidurans CH34
J.MOL.BIOL., 339, 2004
|
|
1OSG
 
 | | Complex between BAFF and a BR3 derived peptide presented in a beta-hairpin scaffold | | Descriptor: | BR3 derived PEPTIDE, MAGNESIUM ION, Tumor necrosis factor ligand superfamily member 13B | | Authors: | Gordon, N.C, Pan, B, Hymowitz, S.G, Yin, J.P, Kelley, R.F, Cochran, A.G, Yan, M, Dixit, V.M, Fairbrother, W.J, Starovasnik, M.A. | | Deposit date: | 2003-03-19 | | Release date: | 2003-05-27 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | BAFF/BLyS receptor 3 comprises a minimal TNF receptor-like module that encodes a highly focused ligand-binding site
Biochemistry, 42, 2003
|
|
1OSI
 
 | | STRUCTURE OF 3-ISOPROPYLMALATE DEHYDROGENASE | | Descriptor: | 3-ISOPROPYLMALATE DEHYDROGENASE | | Authors: | Qu, C, Akanuma, S, Moriyama, H, Tanaka, N, Oshima, T. | | Deposit date: | 1996-10-22 | | Release date: | 1997-01-27 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | A mutation at the interface between domains causes rearrangement of domains in 3-isopropylmalate dehydrogenase.
Protein Eng., 10, 1997
|
|
1OSJ
 
 | | STRUCTURE OF 3-ISOPROPYLMALATE DEHYDROGENASE | | Descriptor: | 3-ISOPROPYLMALATE DEHYDROGENASE | | Authors: | Qu, C, Akanuma, S, Moriyama, H, Tanaka, N, Oshima, T. | | Deposit date: | 1996-10-22 | | Release date: | 1997-01-27 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | A mutation at the interface between domains causes rearrangement of domains in 3-isopropylmalate dehydrogenase.
Protein Eng., 10, 1997
|
|
1OSM
 
 | | OSMOPORIN (OMPK36) FROM KLEBSIELLA PNEUMONIAE | | Descriptor: | DODECANE, OMPK36 | | Authors: | Dutzler, R, Schirmer, T. | | Deposit date: | 1999-01-08 | | Release date: | 1999-07-26 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Crystal structure and functional characterization of OmpK36, the osmoporin of Klebsiella pneumoniae.
Structure Fold.Des., 7, 1999
|
|
1OSN
 
 | | Crystal structure of Varicella zoster virus thymidine kinase in complex with BVDU-MP and ADP | | Descriptor: | (E)-5-(2-BROMOVINYL)-2'-DEOXYURIDINE-5'-MONOPHOSPHATE, ADENOSINE-5'-DIPHOSPHATE, Thymidine kinase | | Authors: | Bird, L.E, Ren, J, Wright, A, Leslie, K.D, Degreve, B, Balzarini, J, Stammers, D.K. | | Deposit date: | 2003-03-20 | | Release date: | 2003-06-10 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Crystal structure of varicella zoster virus thymidine kinase
J.Biol.Chem., 278, 2003
|
|
1OSS
 
 | | T190P STREPTOMYCES GRISEUS TRYPSIN IN COMPLEX WITH BENZAMIDINE | | Descriptor: | BENZAMIDINE, CALCIUM ION, SULFATE ION, ... | | Authors: | Page, M.J, Wong, S.L, Hewitt, J, Strynadka, N.C, MacGillivray, R.T. | | Deposit date: | 2003-03-20 | | Release date: | 2003-08-19 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.93 Å) | | Cite: | Engineering the Primary Substrate Specificity of Streptomyces griseus Trypsin.
Biochemistry, 42, 2003
|
|
1OSU
 
 | |
1OSV
 
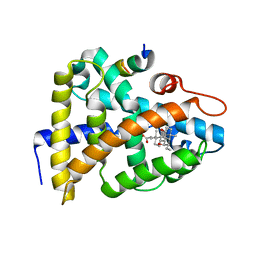 | | STRUCTURAL BASIS FOR BILE ACID BINDING AND ACTIVATION OF THE NUCLEAR RECEPTOR FXR | | Descriptor: | 6-ETHYL-CHENODEOXYCHOLIC ACID, Bile acid receptor, Nuclear receptor coactivator 2 | | Authors: | Mi, L.Z, Devarakonda, S, Harp, J.M, Han, Q, Pellicciari, R, Willson, T.M, Khorasanizadeh, S, Rastinejad, F. | | Deposit date: | 2003-03-20 | | Release date: | 2004-03-23 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structural Basis for Bile Acid Binding and Activation of the Nuclear Receptor FXR
Mol.Cell, 11, 2003
|
|
1OSY
 
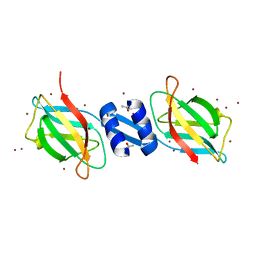 | | Crystal structure of FIP-Fve fungal immunomodulatory protein | | Descriptor: | BROMIDE ION, IMMUNOMODULATORY PROTEIN FIP-FVE | | Authors: | Palasingam, P, Joseph, J.S, Seow, S.V, Shai, V, Robinson, H, Chua, K.Y, Kolatkar, P.R. | | Deposit date: | 2003-03-20 | | Release date: | 2003-11-04 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | A 1.7A structure of Fve, a member of the new fungal
immunomodulatory protein family
J.Mol.Biol., 332, 2003
|
|
1OSZ
 
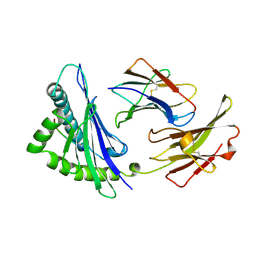 | | MHC CLASS I H-2KB HEAVY CHAIN COMPLEXED WITH BETA-2 MICROGLOBULIN AND AN (L4V) MUTANT OF THE VESICULAR STOMATITIS VIRUS NUCLEOPROTEIN | | Descriptor: | BETA-2 MICROGLOBULIN, MHC CLASS I H-2KB HEAVY CHAIN, VESICULAR STOMATITIS VIRUS NUCLEOPROTEIN | | Authors: | Ghendler, Y, Teng, M.-K, Liu, J.-H, Witte, T, Liu, J, Kim, K.S, Kern, P, Chang, H.-C, Wang, J.-H, Reinherz, E.L. | | Deposit date: | 1998-06-18 | | Release date: | 1999-01-13 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Differential thymic selection outcomes stimulated by focal structural alteration in peptide/major histocompatibility complex ligands.
Proc.Natl.Acad.Sci.USA, 95, 1998
|
|
1OT1
 
 | |
1OT2
 
 | |
1OT3
 
 | | Crystal structure of Drosophila deoxyribonucleotide kinase complexed with the substrate deoxythymidine | | Descriptor: | Deoxyribonucleoside Kinase, SULFATE ION, THYMIDINE | | Authors: | Mikkelsen, N.E, Johansson, K, Karlsson, A, Knecht, W, Andersen, G, Piskur, J, Munch-Petersen, B, Eklund, H. | | Deposit date: | 2003-03-21 | | Release date: | 2003-05-27 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structural Basis for Feedback Inhibition of the Deoxyribonucleoside Salvage Pathway: Studies of the Drosophila Deoxyribonucleoside Kinase.
Biochemistry, 42, 2003
|
|
1OT5
 
 | | The 2.4 Angstrom Crystal Structure of Kex2 in complex with a peptidyl-boronic acid inhibitor | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Ac-Ala-Lys-boroArg N-acetylated boronic acid peptide inhibitor, ... | | Authors: | Holyoak, T, Wilson, M.A, Fenn, T.D, Kettner, C.A, Petsko, G.A, Fuller, R.S, Ringe, D. | | Deposit date: | 2003-03-21 | | Release date: | 2003-06-17 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | 2.4 A Resolution Crystal Structure of the Prototypical Hormone-Processing Protease Kex2 in Complex with an Ala-Lys-Arg Boronic Acid Inhibitor
Biochemistry, 42, 2003
|
|
1OT6
 
 | |
1OT7
 
 | | Structural Basis for 3-deoxy-CDCA Binding and Activation of FXR | | Descriptor: | 6-ETHYL-CHENODEOXYCHOLIC ACID, Bile Acid Receptor, ISO-URSODEOXYCHOLIC ACID, ... | | Authors: | Mi, L.Z, Devarakonda, S, Harp, J.M, Han, Q, Pellicciari, R, Willson, T.M, Khorasanizadeh, S, Rastinejad, F. | | Deposit date: | 2003-03-21 | | Release date: | 2004-03-23 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Structural Basis for Bile Acid Binding and Activation of the Nuclear Receptor FXR
Mol.Cell, 11, 2003
|
|
1OT8
 
 | | Structure of the Ankyrin Domain of the Drosophila Notch Receptor | | Descriptor: | MAGNESIUM ION, Neurogenic locus Notch protein | | Authors: | Zweifel, M.E, Leahy, D.J, Hughson, F.M, Barrick, D. | | Deposit date: | 2003-03-21 | | Release date: | 2003-10-28 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structure and stability of the ankyrin domain of the Drosophila Notch receptor
Protein Sci., 12, 2003
|
|
1OT9
 
 | | CRYOTRAPPED STATE IN WILD TYPE PHOTOACTIVE YELLOW PROTEIN, INDUCED WITH CONTINUOUS ILLUMINATION AT 110K | | Descriptor: | 4'-HYDROXYCINNAMIC ACID, Photoactive yellow protein | | Authors: | Anderson, S, Crosson, S, Moffat, K. | | Deposit date: | 2003-03-21 | | Release date: | 2004-05-11 | | Last modified: | 2019-07-24 | | Method: | X-RAY DIFFRACTION (1 Å) | | Cite: | Short hydrogen bonds in photoactive yellow protein.
Acta Crystallogr.,Sect.D, 60, 2004
|
|
1OTA
 
 | | E46Q MUTANT OF PHOTOACTIVE YELLOW PROTEIN, P63 AT 295K | | Descriptor: | 4'-HYDROXYCINNAMIC ACID, Photoactive yellow protein | | Authors: | Anderson, S, Crosson, S, Moffat, K. | | Deposit date: | 2003-03-21 | | Release date: | 2004-05-11 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (1.1 Å) | | Cite: | Short hydrogen bonds in photoactive yellow protein.
Acta Crystallogr.,Sect.D, 60, 2004
|
|
1OTB
 
 | | WILD TYPE PHOTOACTIVE YELLOW PROTEIN, P63 AT 295K | | Descriptor: | 4'-HYDROXYCINNAMIC ACID, Photoactive yellow protein | | Authors: | Anderson, S, Crosson, S, Moffat, K. | | Deposit date: | 2003-03-21 | | Release date: | 2004-05-11 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (1.1 Å) | | Cite: | Short hydrogen bonds in photoactive yellow protein.
Acta Crystallogr.,Sect.D, 60, 2004
|
|
1OTC
 
 | | THE O. NOVA TELOMERE END BINDING PROTEIN COMPLEXED WITH SINGLE STRAND DNA | | Descriptor: | DNA (5'-D(*GP*GP*GP*GP*TP*TP*TP*TP*GP*GP*GP*G)-3'), PROTEIN (TELOMERE-BINDING PROTEIN ALPHA SUBUNIT), PROTEIN (TELOMERE-BINDING PROTEIN BETA SUBUNIT) | | Authors: | Horvath, M.P, Schweiker, V.L, Bevilacqua, J.M, Ruggles, J.A, Schultz, S.C. | | Deposit date: | 1998-11-25 | | Release date: | 1999-04-12 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Crystal structure of the Oxytricha nova telomere end binding protein complexed with single strand DNA.
Cell(Cambridge,Mass.), 95, 1998
|
|
1OTD
 
 | |
1OTE
 
 | | E46Q MUTANT OF PHOTOACTIVE YELLOW PROTEIN, P65 AT 110K | | Descriptor: | 4'-HYDROXYCINNAMIC ACID, photoactive yellow protein, PYP | | Authors: | Anderson, S, Crosson, S, Moffat, K. | | Deposit date: | 2003-03-21 | | Release date: | 2004-05-11 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Short hydrogen bonds in photoactive yellow protein.
Acta Crystallogr.,Sect.D, 60, 2004
|
|
