1LWY
 
 | | hOgg1 Borohydride-Trapped Intermediate without 8-oxoguanine | | Descriptor: | 5'-D(*GP*CP*GP*TP*CP*CP*AP*(PED)P*GP*TP*CP*TP*AP*CP*C)-3', 5'-D(*GP*GP*TP*AP*GP*AP*CP*CP*TP*GP*GP*AP*CP*GP*C)-3', 8-OXOGUANINE DNA GLYCOSYLASE | | Authors: | Fromme, J.C, Bruner, S.D, Yang, W, Karplus, M, Verdine, G.L. | | Deposit date: | 2002-06-03 | | Release date: | 2003-02-25 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.01 Å) | | Cite: | Product-Assisted Catalysis in Base Excision DNA Repair
Nat.Struct.Biol., 10, 2003
|
|
1LX5
 
 | | Crystal Structure of the BMP7/ActRII Extracellular Domain Complex | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Activin Type II Receptor, alpha-D-mannopyranose-(1-3)-[beta-D-mannopyranose-(1-4)][alpha-D-mannopyranose-(1-6)]beta-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Greenwald, J, Groppe, J, Kwiatkowski, W, Choe, S. | | Deposit date: | 2002-06-04 | | Release date: | 2003-04-01 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (3.3 Å) | | Cite: | The BMP7/ActRII Extracellular Domain Complex Provides New Insights into
the Cooperative Nature of Receptor Assembly
Mol.Cell, 11, 2003
|
|
1LX7
 
 | | Structure of E. coli uridine phosphorylase at 2.0A | | Descriptor: | uridine phosphorylase | | Authors: | Burling, T, Buglino, J.A, Kniewel, R, Chadna, T, Beckwith, A, Lima, C.D, Burley, S.K, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2002-06-04 | | Release date: | 2002-06-12 | | Last modified: | 2021-02-03 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structure of Escherichia coli uridine phosphorylase at 2.0 A.
Acta Crystallogr.,Sect.D, 59, 2003
|
|
1LXD
 
 | | CRYSTAL STRUCTURE OF THE RAS INTERACTING DOMAIN OF RALGDS, A GUANINE NUCLEOTIDE DISSOCIATION STIMULATOR OF RAL PROTEIN | | Descriptor: | RALGDSB | | Authors: | Huang, L, Weng, X.W, Hofer, F, Martin, G.S, Kim, S.H. | | Deposit date: | 1997-03-05 | | Release date: | 1998-03-11 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Three-dimensional structure of the Ras-interacting domain of RalGDS.
Nat.Struct.Biol., 4, 1997
|
|
1LXE
 
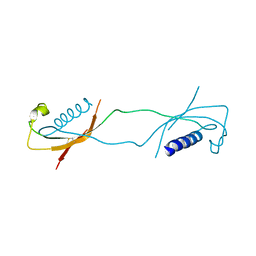 | | CRYSTAL STRUCTURE OF THE CATHELICIDIN MOTIF OF PROTEGRINS | | Descriptor: | protegrin-3 precursor | | Authors: | Sanchez, J.F, Hoh, F, Strub, M.P, Aumelas, A, Dumas, C. | | Deposit date: | 2002-06-05 | | Release date: | 2002-10-09 | | Last modified: | 2017-10-11 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structure of the cathelicidin motif of protegrin-3 precursor: structural insights into the activation mechanism of an antimicrobial protein.
Structure, 10, 2002
|
|
1LXI
 
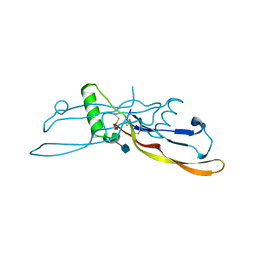 | | Refinement of BMP7 crystal structure | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, BONE MORPHOGENETIC PROTEIN 7 | | Authors: | Greenwald, J, Groppe, J, Kwiatkowski, W, Choe, S. | | Deposit date: | 2002-06-05 | | Release date: | 2003-04-01 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | The BMP7/ActRII Extracellular Domain Complex Provides New Insights into
the Cooperative Nature of Receptor Assembly
Mol.Cell, 11, 2003
|
|
1LXJ
 
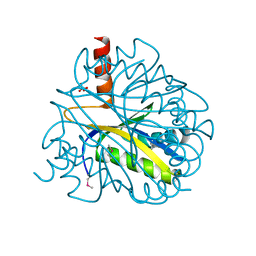 | | X-RAY STRUCTURE OF YBL001c NORTHEAST STRUCTURAL GENOMICS (NESG) CONSORTIUM TARGET YTYst72 | | Descriptor: | HYPOTHETICAL 11.5KDA PROTEIN IN HTB2-NTH2 INTERGENIC REGION, SULFATE ION | | Authors: | Tao, X, Khayat, R, Christendat, D, Savchenko, A, Xu, X, Edwards, A, Arrowsmith, C.H, Tong, L, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2002-06-05 | | Release date: | 2003-07-29 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | CRYSTAL STRUCTURES OF MTH1187 AND ITS YEAST ORTHOLOG YBL001C
Proteins, 52, 2003
|
|
1LXK
 
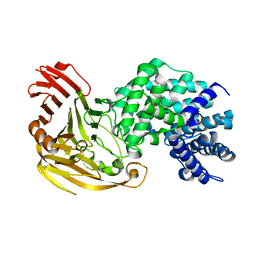 | | Streptococcus pneumoniae Hyaluronate Lyase in Complex with Tetrasaccharide Hyaluronan Substrate | | Descriptor: | Hyaluronate Lyase, beta-D-glucopyranuronic acid-(1-3)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-beta-D-glucopyranuronic acid-(1-3)-2-acetamido-2-deoxy-beta-D-glucopyranose | | Authors: | Jedrzejas, M.J, Mello, L.V, De Groot, B.L, Li, S. | | Deposit date: | 2002-06-05 | | Release date: | 2002-08-07 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.53 Å) | | Cite: | Mechanism of hyaluronan degradation by Streptococcus pneumoniae hyaluronate lyase. Structures of complexes with the substrate.
J.Biol.Chem., 277, 2002
|
|
1LXM
 
 | | Crystal Structure of Streptococcus agalactiae Hyaluronate Lyase Complexed with Hexasaccharide Unit of Hyaluronan | | Descriptor: | HYALURONATE Lyase, beta-D-glucopyranuronic acid-(1-3)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-beta-D-glucopyranuronic acid-(1-3)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-beta-D-glucopyranuronic acid-(1-3)-2-acetamido-2-deoxy-beta-D-glucopyranose | | Authors: | Mello, L.V, de Groot, B.L, Li, S, Jedrzejas, M.J. | | Deposit date: | 2002-06-05 | | Release date: | 2002-10-30 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structure and flexibility of Streptococcus agalactiae hyaluronate lyase complex with its substrate. Insights into the mechanism of processive degradation of hyaluronan.
J.Biol.Chem., 277, 2002
|
|
1LXN
 
 | | X-RAY STRUCTURE OF MTH1187 NORTHEAST STRUCTURAL GENOMICS CONSORTIUM TARGET TT272 | | Descriptor: | HYPOTHETICAL PROTEIN MTH1187, SULFATE ION | | Authors: | Tao, X, Khayat, R, Christendat, D, Savchenko, A, Xu, X, Edwards, A, Arrowsmith, C.H, Tong, L, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2002-06-05 | | Release date: | 2003-07-29 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystal Structures of MTH1187 and its Yeast Ortholog YBL001C
Proteins, 52, 2003
|
|
1LXT
 
 | | STRUCTURE OF PHOSPHOTRANSFERASE PHOSPHOGLUCOMUTASE FROM RABBIT | | Descriptor: | CADMIUM ION, PHOSPHOGLUCOMUTASE (DEPHOSPHO FORM), SULFATE ION | | Authors: | Ray Junior, W.J, Baranidharan, S, Liu, Y. | | Deposit date: | 1996-07-28 | | Release date: | 1997-02-12 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Structure of rabbit muscle phosphoglucomutase refined at 2.4 A resolution.
Acta Crystallogr.,Sect.D, 53, 1997
|
|
1LXY
 
 | | Crystal Structure of Arginine Deiminase covalently linked with L-citrulline | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, Arginine Deiminase, CITRULLINE | | Authors: | Das, K, Buttler, G.H, Kwiatkowski, V, Yadav, P, Arnold, E. | | Deposit date: | 2002-06-06 | | Release date: | 2004-01-20 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal structures of arginine deiminase with covalent reaction intermediates; implications for catalytic mechanism
Structure, 12, 2004
|
|
1LXZ
 
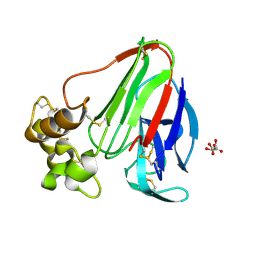 | | Structure of thaumatin crystallized in the presence of glycerol | | Descriptor: | L(+)-TARTARIC ACID, Thaumatin I | | Authors: | Charron, C, Kadri, A, Robert, M.C, Giege, R, Lorber, B. | | Deposit date: | 2002-06-06 | | Release date: | 2003-01-14 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.25 Å) | | Cite: | Crystallization in the presence of glycerol displaces water molecules in the structure of thaumatin.
Acta Crystallogr.,Sect.D, 58, 2002
|
|
1LY0
 
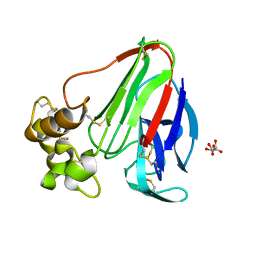 | | Structure of thaumatin crystallized in the presence of glycerol | | Descriptor: | L(+)-TARTARIC ACID, Thaumatin I | | Authors: | Charron, C, Kadri, A, Robert, M.C, Giege, R, Lorber, B. | | Deposit date: | 2002-06-06 | | Release date: | 2003-01-14 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.36 Å) | | Cite: | Crystallization in the presence of glycerol displaces water molecules in the structure of thaumatin.
Acta Crystallogr.,Sect.D, 58, 2002
|
|
1LY1
 
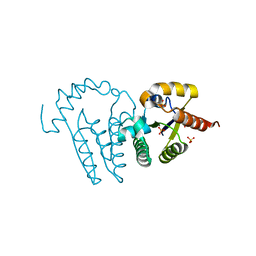 | |
1LY2
 
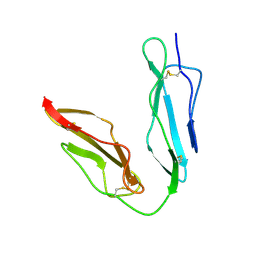 | | Crystal structure of unliganded human CD21 SCR1-SCR2 (Complement receptor type 2) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, complement receptor type 2 | | Authors: | Prota, A.E, Sage, D.R, Stehle, T, Fingeroth, J.D. | | Deposit date: | 2002-06-06 | | Release date: | 2002-07-05 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | The crystal structure of human CD21: Implications for Epstein-Barr virus and C3d binding.
Proc.Natl.Acad.Sci.USA, 99, 2002
|
|
1LY3
 
 | | ANALYSIS OF QUINAZOLINE AND PYRIDOPYRIMIDINE N9-C10 REVERSED BRIDGE ANTIFOLATES IN COMPLEX WITH NADP+ AND PNEUMOCYSTIS CARINII DIHYDROFOLATE REDUCTASE | | Descriptor: | 2,4-DIAMINO-6-[N-(2',5'-DIMETHOXYBENZYL)-N-METHYLAMINO]QUINAZOLINE, DIHYDROFOLATE REDUCTASE, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE | | Authors: | Cody, V, Galitsky, N, Luft, J.R, Pangborn, W, Queener, S.F, Gangjee, A. | | Deposit date: | 2002-06-06 | | Release date: | 2002-08-28 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Analysis of quinazoline and pyrido[2,3-d]pyrimidine N9-C10 reversed-bridge antifolates in complex with NADP+ and Pneumocystis carinii dihydrofolate reductase.
Acta Crystallogr.,Sect.D, 58, 2002
|
|
1LY4
 
 | | Analysis of quinazoline and PYRIDO[2,3D]PYRIMIDINE N9-C10 reversed bridge antifolates in complex with NADP+ and Pneumocystis carinii dihydrofolate reductase | | Descriptor: | 2,4-DIAMINO-6-[N-(3',5'-DIMETHOXYBENZYL)-N-METHYLAMINO]PYRIDO[2,3-D]PYRIMIDINE, DIHYDROFOLATE REDUCTASE, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE | | Authors: | Cody, V, Galitsky, N, Luft, J.R, Pangborn, W, Queener, S.F, Gangjee, A. | | Deposit date: | 2002-06-06 | | Release date: | 2002-08-28 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Analysis of quinazoline and pyrido[2,3-d]pyrimidine N9-C10 reversed-bridge antifolates in complex with NADP+ and Pneumocystis carinii dihydrofolate reductase.
Acta Crystallogr.,Sect.D, 58, 2002
|
|
1LY8
 
 | | The crystal structure of a mutant enzyme of Coprinus cinereus peroxidase provides an understanding of its increased thermostability and insight into modelling of protein structures | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, GLYCEROL, ... | | Authors: | Houborg, K, Harris, P, Poulsen, J.-C.N, Svendsen, A, Schneider, P, Larsen, S. | | Deposit date: | 2002-06-07 | | Release date: | 2002-06-14 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | The structure of a mutant enzyme of Coprinus cinereus peroxidase provides an understanding of its increased thermostability.
Acta Crystallogr.,Sect.D, 59, 2003
|
|
1LY9
 
 | | The impact of the physical and chemical environment on the molecular structure of Coprinus cinereus peroxidase | | Descriptor: | CALCIUM ION, PROTOPORPHYRIN IX CONTAINING FE, Peroxidase | | Authors: | Houborg, K, Harris, P, Petersen, J.F.W, Rowland, P, Poulsen, J.-C.N, Schneider, P, Vind, J, Larsen, S. | | Deposit date: | 2002-06-07 | | Release date: | 2002-06-14 | | Last modified: | 2021-10-27 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Impact of the physical and chemical environment on the molecular structure of Coprinus cinereus peroxidase.
Acta Crystallogr.,Sect.D, 59, 2003
|
|
1LYC
 
 | | The impact of the physical and chemical enviroment on the molecular structure of Coprinus cinereus peroxidase | | Descriptor: | CALCIUM ION, PROTOPORPHYRIN IX CONTAINING FE, Peroxidase | | Authors: | Houborg, K, Harris, P, Petersen, J.F.W, Rowland, P, Poulsen, J.-C.N, Schneider, P, Vind, J, Larsen, S. | | Deposit date: | 2002-06-07 | | Release date: | 2002-06-14 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.57 Å) | | Cite: | Impact of the physical and chemical environment on the molecular structure of Coprinus cinereus peroxidase.
Acta Crystallogr.,Sect.D, 59, 2003
|
|
1LYE
 
 | | DISSECTION OF HELIX CAPPING IN T4 LYSOZYME BY STRUCTURAL AND THERMODYNAMIC ANALYSIS OF SIX AMINO ACID SUBSTITUTIONS AT THR 59 | | Descriptor: | BETA-MERCAPTOETHANOL, CHLORIDE ION, T4 LYSOZYME | | Authors: | Bell, J.A, Becktel, W.J, Sauer, U, Baase, W.A, Matthews, B.W. | | Deposit date: | 1992-08-10 | | Release date: | 1993-10-31 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Dissection of helix capping in T4 lysozyme by structural and thermodynamic analysis of six amino acid substitutions at Thr 59.
Biochemistry, 31, 1992
|
|
1LYF
 
 | | DISSECTION OF HELIX CAPPING IN T4 LYSOZYME BY STRUCTURAL AND THERMODYNAMIC ANALYSIS OF SIX AMINO ACID SUBSTITUTIONS AT THR 59 | | Descriptor: | BETA-MERCAPTOETHANOL, CHLORIDE ION, T4 LYSOZYME | | Authors: | Bell, J.A, Becktel, W.J, Sauer, U, Baase, W.A, Matthews, B.W. | | Deposit date: | 1992-08-10 | | Release date: | 1993-10-31 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Dissection of helix capping in T4 lysozyme by structural and thermodynamic analysis of six amino acid substitutions at Thr 59.
Biochemistry, 31, 1992
|
|
1LYG
 
 | | DISSECTION OF HELIX CAPPING IN T4 LYSOZYME BY STRUCTURAL AND THERMODYNAMIC ANALYSIS OF SIX AMINO ACID SUBSTITUTIONS AT THR 59 | | Descriptor: | BETA-MERCAPTOETHANOL, CHLORIDE ION, T4 LYSOZYME | | Authors: | Bell, J.A, Becktel, W.J, Sauer, U, Baase, W.A, Matthews, B.W. | | Deposit date: | 1992-08-10 | | Release date: | 1993-10-31 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Dissection of helix capping in T4 lysozyme by structural and thermodynamic analysis of six amino acid substitutions at Thr 59.
Biochemistry, 31, 1992
|
|
1LYH
 
 | | DISSECTION OF HELIX CAPPING IN T4 LYSOZYME BY STRUCTURAL AND THERMODYNAMIC ANALYSIS OF SIX AMINO ACID SUBSTITUTIONS AT THR 59 | | Descriptor: | BETA-MERCAPTOETHANOL, CHLORIDE ION, T4 LYSOZYME | | Authors: | Bell, J.A, Becktel, W.J, Sauer, U, Baase, W.A, Matthews, B.W. | | Deposit date: | 1992-08-10 | | Release date: | 1993-10-31 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Dissection of helix capping in T4 lysozyme by structural and thermodynamic analysis of six amino acid substitutions at Thr 59.
Biochemistry, 31, 1992
|
|
