1IJE
 
 | | Nucleotide Exchange Intermediates in the eEF1A-eEF1Ba Complex | | Descriptor: | GUANOSINE-5'-DIPHOSPHATE, elongation factor 1-alpha, elongation factor 1-beta | | Authors: | Andersen, G.R, Valente, L, Pedersen, L, Kinzy, T.G, Nyborg, J. | | Deposit date: | 2001-04-26 | | Release date: | 2001-05-09 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Crystal structures of nucleotide exchange intermediates in the eEF1A-eEF1Balpha complex.
Nat.Struct.Biol., 8, 2001
|
|
1IJF
 
 | | Nucleotide exchange mechanisms in the eEF1A-eEF1Ba complex | | Descriptor: | GUANOSINE-5'-DIPHOSPHATE, MAGNESIUM ION, elongation factor 1-alpha, ... | | Authors: | Andersen, G.R, Valente, L, Pedersen, L, Kinzy, T.G, Nyborg, J. | | Deposit date: | 2001-04-26 | | Release date: | 2001-05-09 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Crystal structures of nucleotide exchange intermediates in the eEF1A-eEF1Balpha complex.
Nat.Struct.Biol., 8, 2001
|
|
1IJG
 
 | | Structure of the Bacteriophage phi29 Head-Tail Connector Protein | | Descriptor: | UPPER COLLAR PROTEIN | | Authors: | Simpson, A.A, Leiman, P.G, Tao, Y, He, Y, Badasso, M, Jardine, P.J, Anderson, D.L, Rossmann, M.G. | | Deposit date: | 2001-04-26 | | Release date: | 2001-05-09 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Structure determination of the head-tail connector of bacteriophage phi29.
Acta Crystallogr.,Sect.D, 57, 2001
|
|
1IJH
 
 | | CHOLESTEROL OXIDASE FROM STREPTOMYCES ASN485LEU MUTANT | | Descriptor: | CHOLESTEROL OXIDASE, FLAVIN-ADENINE DINUCLEOTIDE | | Authors: | Vrielink, A, Lario, P.I. | | Deposit date: | 2001-04-26 | | Release date: | 2001-12-28 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (1.53 Å) | | Cite: | The presence of a hydrogen bond between asparagine 485 and the pi system of FAD modulates the redox potential in the reaction catalyzed by cholesterol oxidase.
Biochemistry, 40, 2001
|
|
1IJI
 
 | | Crystal Structure of L-Histidinol Phosphate Aminotransferase with PLP | | Descriptor: | Histidinol Phosphate Aminotransferase, PYRIDOXAL-5'-PHOSPHATE | | Authors: | Sivaraman, J, Li, Y, Larocque, R, Schrag, J.D, Cygler, M, Matte, A. | | Deposit date: | 2001-04-26 | | Release date: | 2001-08-29 | | Last modified: | 2017-10-04 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystal structure of histidinol phosphate aminotransferase (HisC) from Escherichia coli, and its covalent complex with pyridoxal-5'-phosphate and l-histidinol phosphate.
J.Mol.Biol., 311, 2001
|
|
1IJK
 
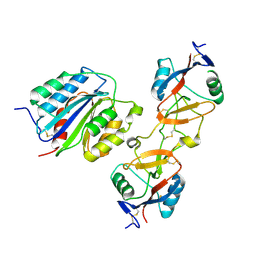 | | The von Willebrand Factor mutant (I546V) A1 domain-botrocetin Complex | | Descriptor: | Botrocetin, von Willebrand factor | | Authors: | Fukuda, K, Doggett, T.A, Bankston, L.A, Cruz, M.A, Diacovo, T.G, Liddington, R.C. | | Deposit date: | 2001-04-26 | | Release date: | 2002-07-10 | | Last modified: | 2021-10-27 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Structural basis of von Willebrand factor activation by the snake toxin botrocetin.
Structure, 10, 2002
|
|
1IJL
 
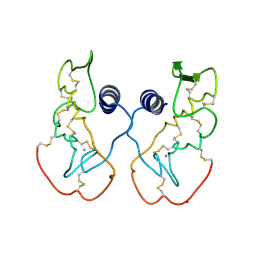 | | Crystal structure of acidic phospholipase A2 from deinagkistrodon acutus | | Descriptor: | CALCIUM ION, PHOSPHOLIPASE A2, ZINC ION | | Authors: | Gu, L, Zhang, H, Song, S, Zhou, Y, Lin, Z. | | Deposit date: | 2001-04-27 | | Release date: | 2001-12-28 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Structure of an acidic phospholipase A2 from the venom of Deinagkistrodon acutus.
Acta Crystallogr.,Sect.D, 58, 2002
|
|
1IJQ
 
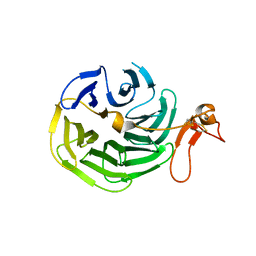 | | Crystal Structure of the LDL Receptor YWTD-EGF Domain Pair | | Descriptor: | LOW-DENSITY LIPOPROTEIN RECEPTOR | | Authors: | Jeon, H, Meng, W, Takagi, J, Eck, M.J, Springer, T.A, Blacklow, S.C. | | Deposit date: | 2001-04-27 | | Release date: | 2001-05-23 | | Last modified: | 2018-04-04 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Implications for familial hypercholesterolemia from the structure of the LDL receptor YWTD-EGF domain pair.
Nat.Struct.Biol., 8, 2001
|
|
1IJS
 
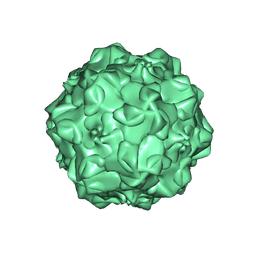 | | CPV (STRAIN D) mutant A300D, complex (VIRAL COAT/DNA), VP2, PH=7.5, T=4 DEGREES C | | Descriptor: | DNA (5'-D(*AP*C)-3'), DNA (5'-D(*CP*CP*AP*CP*CP*CP*CP*AP*A)-3'), PROTEIN (PARVOVIRUS COAT PROTEIN) | | Authors: | Llamas-Saiz, A.L, Agbandje-McKenna, M, Parker, J.S.L, Wahid, A.T.M, Parrish, C.R, Rossmann, M.G. | | Deposit date: | 1996-09-12 | | Release date: | 1996-12-23 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (3.25 Å) | | Cite: | Structural analysis of a mutation in canine parvovirus which controls antigenicity and host range.
Virology, 225, 1996
|
|
1IJU
 
 | | HUMAN BETA-DEFENSIN-1 | | Descriptor: | BETA-DEFENSIN 1, GLYCEROL, SULFATE ION | | Authors: | Hoover, D.M, Lubkowski, J. | | Deposit date: | 2001-04-30 | | Release date: | 2001-10-24 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | The structure of human beta-defensin-1: new insights into structural properties of beta-defensins.
J.Biol.Chem., 276, 2001
|
|
1IJV
 
 | | HUMAN BETA-DEFENSIN-1 | | Descriptor: | BROMIDE ION, Beta-defensin 1, POTASSIUM ION, ... | | Authors: | Hoover, D.M, Lubkowski, J. | | Deposit date: | 2001-04-30 | | Release date: | 2001-10-24 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.2 Å) | | Cite: | The structure of human beta-defensin-1: new insights into structural properties of beta-defensins.
J.Biol.Chem., 276, 2001
|
|
1IJW
 
 | | Testing the Water-Mediated Hin Recombinase DNA Recognition by Systematic Mutations. | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, 5'-D(*AP*TP*(CBR)P*TP*TP*AP*TP*CP*AP*AP*AP*AP*AP*C)-3', 5'-D(*TP*GP*TP*TP*TP*TP*TP*GP*AP*TP*AP*AP*GP*A)-3', ... | | Authors: | Chiu, T.K, Sohn, C, Johnson, R.C, Dickerson, R.E. | | Deposit date: | 2001-04-30 | | Release date: | 2002-02-22 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Testing water-mediated DNA recognition by the Hin recombinase.
EMBO J., 21, 2002
|
|
1IJX
 
 | | CRYSTAL STRUCTURE OF THE CYSTEINE-RICH DOMAIN OF SECRETED FRIZZLED-RELATED PROTEIN 3 (SFRP-3;FZB) | | Descriptor: | SECRETED FRIZZLED-RELATED SEQUENCE PROTEIN 3, SULFATE ION | | Authors: | Dann III, C.E, Hsieh, J.C, Rattner, A, Sharma, D, Nathans, J, Leahy, D.J. | | Deposit date: | 2001-04-30 | | Release date: | 2001-07-11 | | Last modified: | 2021-10-27 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Insights into Wnt binding and signalling from the structures of two Frizzled cysteine-rich domains.
Nature, 412, 2001
|
|
1IJY
 
 | | CRYSTAL STRUCTURE OF THE CYSTEINE-RICH DOMAIN OF MOUSE FRIZZLED 8 (MFZ8) | | Descriptor: | FRIZZLED HOMOLOG 8 | | Authors: | Dann III, C.E, Hsieh, J.C, Rattner, A, Sharma, D, Nathans, J, Leahy, D.J. | | Deposit date: | 2001-05-01 | | Release date: | 2001-07-11 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (1.35 Å) | | Cite: | Insights into Wnt binding and signalling from the structures of two Frizzled cysteine-rich domains.
Nature, 412, 2001
|
|
1IK3
 
 | | LIPOXYGENASE-3 (SOYBEAN) COMPLEX WITH 13(S)-HYDROPEROXY-9(Z),11(E)-OCTADECADIENOIC ACID | | Descriptor: | (TRANS-12,13-EPOXY)-11-HYDROXY-9(Z)-OCTADECENOIC ACID, (TRANS-12,13-EPOXY)-9-HYDROXY-10(E)-OCTADECENOIC ACID, 13(R)-HYDROPEROXY-9(Z),11(E)-OCTADECADIENOIC ACID, ... | | Authors: | Skrzypczak-Jankun, E, Funk Jr, M.O. | | Deposit date: | 2001-05-01 | | Release date: | 2001-11-14 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Three-dimensional structure of a purple lipoxygenase.
J.Am.Chem.Soc., 123, 2001
|
|
1IK4
 
 | | X-ray Structure of Methylglyoxal Synthase from E. coli Complexed with Phosphoglycolohydroxamic Acid | | Descriptor: | METHYLGLYOXAL SYNTHASE, PHOSPHOGLYCOLOHYDROXAMIC ACID | | Authors: | Marks, G.T, Harris, T.K, Massiah, M.A, Mildvan, A.S, Harrison, D.H.T. | | Deposit date: | 2001-05-02 | | Release date: | 2001-09-26 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Mechanistic implications of methylglyoxal synthase complexed with phosphoglycolohydroxamic acid as observed by X-ray crystallography and NMR spectroscopy.
Biochemistry, 40, 2001
|
|
1IK5
 
 | | Crystal Structure of a 14mer RNA Containing Double UU Bulges in Two Crystal Forms: A Novel U*(AU) Intramolecular Base Triple | | Descriptor: | 5'-R(*GP*GP*UP*AP*UP*UP*UP*CP*GP*GP*UP*AP*(CBR)P*C)-3', 5'-R(*GP*GP*UP*AP*UP*UP*UP*UP*GP*GP*UP*AP*(CBR)P*C)-3', MAGNESIUM ION | | Authors: | Deng, J, Xiong, Y, Sudarsanakumar, C, Shi, K, Sundaralingam, M. | | Deposit date: | 2001-05-02 | | Release date: | 2001-11-09 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal structures of two forms of a 14-mer RNA/DNA chimer duplex with double UU bulges: a novel intramolecular U*(A x U) base triple.
RNA, 7, 2001
|
|
1IK6
 
 | |
1IK7
 
 | | Crystal Structure of the Uncomplexed Pelle Death Domain | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, PROBABLE SERINE/THREONINE-PROTEIN KINASE Pelle | | Authors: | Xiao, T, Gardner, K.H, Sprang, S.R. | | Deposit date: | 2001-05-02 | | Release date: | 2002-07-31 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Cosolvent-induced transformation
of a death domain tertiary structure
Proc.Natl.Acad.Sci.USA, 99, 2002
|
|
1IK9
 
 | | CRYSTAL STRUCTURE OF A XRCC4-DNA LIGASE IV COMPLEX | | Descriptor: | DNA LIGASE IV, DNA REPAIR PROTEIN XRCC4 | | Authors: | Sibanda, B.L, Critchlow, S.E, Begun, J, Pei, X.Y, Jackson, S.P, Blundell, T.L, Pellegrini, L. | | Deposit date: | 2001-05-03 | | Release date: | 2001-11-21 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystal structure of an Xrcc4-DNA ligase IV complex.
Nat.Struct.Biol., 8, 2001
|
|
1IKE
 
 | | Crystal Structure of Nitrophorin 4 from Rhodnius Prolixus Complexed with Histamine at 1.5 A Resolution | | Descriptor: | HISTAMINE, Nitrophorin 4, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Roberts, S.A, Weichsel, A, Qiu, Y, Shelnutt, J.A, Walker, F.A, Montfort, W.R. | | Deposit date: | 2001-05-03 | | Release date: | 2001-10-03 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Ligand-induced heme ruffling and bent no geometry in ultra-high-resolution structures of nitrophorin 4.
Biochemistry, 40, 2001
|
|
1IKF
 
 | | A CONFORMATION OF CYCLOSPORIN A IN AQUEOUS ENVIRONMENT REVEALED BY THE X-RAY STRUCTURE OF A CYCLOSPORIN-FAB COMPLEX | | Descriptor: | CYCLOSPORIN A, IGG1-KAPPA R45-45-11 FAB (HEAVY CHAIN), IGG1-KAPPA R45-45-11 FAB (LIGHT CHAIN) | | Authors: | Vix, O, Altschuh, D, Rees, B, Thierry, J.-C. | | Deposit date: | 1993-12-09 | | Release date: | 1995-03-31 | | Last modified: | 2024-06-05 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | A Conformation of Cyclosporin a in Aqueous Environment Revealed by the X-Ray Structure of a Cyclosporin-Fab Complex.
Science, 256, 1992
|
|
1IKJ
 
 | | 1.27 A CRYSTAL STRUCTURE OF NITROPHORIN 4 FROM RHODNIUS PROLIXUS COMPLEXED WITH IMIDAZOLE | | Descriptor: | IMIDAZOLE, NITROPHORIN 4, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Roberts, S.A, Weichsel, A, Qui, Y, Shelnutt, J.A, Walker, F.A, Montfort, W.R. | | Deposit date: | 2001-05-03 | | Release date: | 2001-10-03 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (1.27 Å) | | Cite: | Ligand-induced heme ruffling and bent no geometry in ultra-high-resolution structures of nitrophorin 4.
Biochemistry, 40, 2001
|
|
1IKK
 
 | |
1IKN
 
 | | IKAPPABALPHA/NF-KAPPAB COMPLEX | | Descriptor: | PROTEIN (I-KAPPA-B-ALPHA), PROTEIN (NF-KAPPA-B P50D SUBUNIT), PROTEIN (NF-KAPPA-B P65 SUBUNIT) | | Authors: | Huxford, T, Huang, D.-B, Malek, S, Ghosh, G. | | Deposit date: | 1998-11-13 | | Release date: | 1999-04-12 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | The crystal structure of the IkappaBalpha/NF-kappaB complex reveals mechanisms of NF-kappaB inactivation.
Cell(Cambridge,Mass.), 95, 1998
|
|
