1RR6
 
 | | Structure of human purine nucleoside phosphorylase in complex with Immucillin-H and phosphate | | Descriptor: | 1,4-DIDEOXY-4-AZA-1-(S)-(9-DEAZAHYPOXANTHIN-9-YL)-D-RIBITOL, PHOSPHATE ION, Purine nucleoside phosphorylase | | Authors: | Shi, W, Lewandowicz, A, Tyler, P.C, Furneaux, R.H, Almo, S.C, Schramm, V.L. | | Deposit date: | 2003-12-08 | | Release date: | 2005-02-22 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Plasmodium falciparum purine nucleoside phosphorylase: crystal structures, immucillin inhibitors, and dual catalytic function.
J.Biol.Chem., 279, 2004
|
|
1RR9
 
 | | Catalytic domain of E.coli Lon protease | | Descriptor: | ATP-dependent protease La, SULFATE ION | | Authors: | Botos, I, Melnikov, E.E, Cherry, S, Tropea, J.E, Khalatova, A.G, Dauter, Z, Maurizi, M.R, Rotanova, T.V, Wlodawer, A, Gustchina, A. | | Deposit date: | 2003-12-08 | | Release date: | 2003-12-23 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | The catalytic domain of Escherichia coli Lon protease has a unique fold and a Ser-Lys dyad in the active site
J.Biol.Chem., 279, 2004
|
|
1RRC
 
 | | T4 POLYNUCLEOTIDE KINASE BOUND TO 5'-GTC-3' SSDNA | | Descriptor: | 5'-D(*GP*TP*C)-3', ADENOSINE-5'-DIPHOSPHATE, CALCIUM ION, ... | | Authors: | Eastberg, J.H, Pelletier, J, Stoddard, B.L. | | Deposit date: | 2003-12-08 | | Release date: | 2004-02-17 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.46 Å) | | Cite: | Recognition of DNA substrates by T4 bacteriophage polynucleotide kinase.
Nucleic Acids Res., 32, 2004
|
|
1RRE
 
 | | Crystal structure of E.coli Lon proteolytic domain | | Descriptor: | ATP-dependent protease La, SULFATE ION | | Authors: | Botos, I, Melnikov, E.E, Cherry, S, Tropea, J.E, Khalatova, A.G, Rasulova, F, Dauter, Z, Maurizi, M.R, Rotanova, T.V, Wlodawer, A, Gustchina, A. | | Deposit date: | 2003-12-08 | | Release date: | 2004-02-03 | | Last modified: | 2021-10-27 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | The catalytic domain of Escherichia coli Lon protease has a unique fold and a Ser-Lys dyad in the active site
J.Biol.Chem., 279, 2004
|
|
1RRF
 
 | | NON-MYRISTOYLATED RAT ADP-RIBOSYLATION FACTOR-1 COMPLEXED WITH GDP, MONOMERIC CRYSTAL FORM | | Descriptor: | GUANOSINE-5'-DIPHOSPHATE, MAGNESIUM ION, RAT ADP-RIBOSYLATION FACTOR-1 | | Authors: | Greasley, S.E, Jhoti, H, Bax, B. | | Deposit date: | 1995-12-16 | | Release date: | 1996-06-20 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | The structure of rat ADP-ribosylation factor-1 (ARF-1) complexed to GDP determined from two different crystal forms.
Nat.Struct.Biol., 2, 1995
|
|
1RRG
 
 | | NON-MYRISTOYLATED RAT ADP-RIBOSYLATION FACTOR-1 COMPLEXED WITH GDP, DIMERIC CRYSTAL FORM | | Descriptor: | GUANOSINE-5'-DIPHOSPHATE, MAGNESIUM ION, RAT ADP-RIBOSYLATION FACTOR-1 | | Authors: | Greasley, S.E, Jhoti, H, Bax, B. | | Deposit date: | 1995-12-16 | | Release date: | 1996-06-20 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | The structure of rat ADP-ribosylation factor-1 (ARF-1) complexed to GDP determined from two different crystal forms.
Nat.Struct.Biol., 2, 1995
|
|
1RRH
 
 | |
1RRK
 
 | | Crystal Structure Analysis of the Bb segment of Factor B | | Descriptor: | COBALT (II) ION, Complement factor B, IODIDE ION, ... | | Authors: | Ponnuraj, K, Xu, Y, Macon, K, Moore, D, Volanakis, J.E, Narayana, S.V. | | Deposit date: | 2003-12-08 | | Release date: | 2004-12-14 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural analysis of engineered Bb fragment of complement factor B: insights into the activation mechanism of the alternative pathway C3-convertase.
Mol.Cell, 14, 2004
|
|
1RRL
 
 | |
1RRO
 
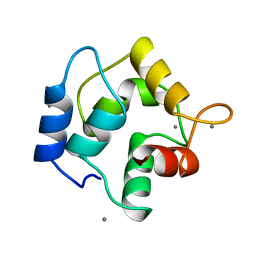 | | REFINEMENT OF RECOMBINANT ONCOMODULIN AT 1.30 ANGSTROMS RESOLUTION | | Descriptor: | CALCIUM ION, RAT ONCOMODULIN | | Authors: | Ahmed, F.R, Rose, D.R, Evans, S.V, Pippy, M.E, To, R. | | Deposit date: | 1992-08-27 | | Release date: | 1993-10-31 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | Refinement of recombinant oncomodulin at 1.30 A resolution.
J.Mol.Biol., 230, 1993
|
|
1RRP
 
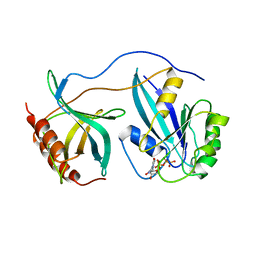 | | STRUCTURE OF THE RAN-GPPNHP-RANBD1 COMPLEX | | Descriptor: | MAGNESIUM ION, NUCLEAR PORE COMPLEX PROTEIN NUP358, PHOSPHOAMINOPHOSPHONIC ACID-GUANYLATE ESTER, ... | | Authors: | Vetter, I.R, Nowak, C, Nishimoto, T, Kuhlmann, J, Wittinghofer, A. | | Deposit date: | 1999-01-15 | | Release date: | 1999-05-18 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.96 Å) | | Cite: | Structure of a Ran-binding domain complexed with Ran bound to a GTP analogue: implications for nuclear transport.
Nature, 398, 1999
|
|
1RRQ
 
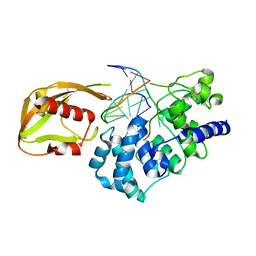 | | MutY adenine glycosylase in complex with DNA containing an A:oxoG pair | | Descriptor: | 5'-D(*TP*GP*TP*CP*CP*AP*AP*GP*TP*CP*T)-3', 5'-D(AP*AP*GP*AP*CP*(8OG)P*TP*GP*GP*AP*C)-3', CALCIUM ION, ... | | Authors: | Fromme, J.C, Banerjee, A, Huang, S.J, Verdine, G.L. | | Deposit date: | 2003-12-08 | | Release date: | 2004-02-17 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.22 Å) | | Cite: | Structural basis for removal of adenine mispaired with 8-oxoguanine by MutY adenine DNA glycosylase
Nature, 427, 2004
|
|
1RRS
 
 | | MutY adenine glycosylase in complex with DNA containing an abasic site | | Descriptor: | 5'-D(*AP*AP*GP*AP*CP*(8OG)P*TP*GP*GP*AP*C)-3', 5'-D(*TP*GP*TP*CP*CP*AP*(HPD)P*GP*TP*CP*T)-3', CALCIUM ION, ... | | Authors: | Fromme, J.C, Banerjee, A, Huang, S.J, Verdine, G.L. | | Deposit date: | 2003-12-08 | | Release date: | 2004-02-17 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structural basis for removal of adenine mispaired with 8-oxoguanine by MutY adenine DNA glycosylase
Nature, 427, 2004
|
|
1RRV
 
 | | X-ray crystal structure of TDP-vancosaminyltransferase GtfD as a complex with TDP and the natural substrate, desvancosaminyl vancomycin. | | Descriptor: | DESVANCOSAMINYL VANCOMYCIN, GLYCEROL, GLYCOSYLTRANSFERASE GTFD, ... | | Authors: | Mulichak, A.M, Lu, W, Losey, H.C, Walsh, C.T, Garavito, R.M. | | Deposit date: | 2003-12-09 | | Release date: | 2004-05-18 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal Structure of Vancosaminyltransferase Gtfd from the Vancomycin Biosynthetic Pathway: Interactions with Acceptor and Nucleotide Ligands
Biochemistry, 43, 2004
|
|
1RS0
 
 | | Crystal Structure Analysis of the Bb segment of Factor B complexed with Di-isopropyl-phosphate (DIP) | | Descriptor: | Complement factor B, DIISOPROPYL PHOSPHONATE, IODIDE ION, ... | | Authors: | Ponnuraj, K, Xu, Y, Macon, K, Moore, D, Volanakis, J.E, Narayana, S.V. | | Deposit date: | 2003-12-09 | | Release date: | 2004-12-14 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Structural analysis of engineered Bb fragment of complement factor B: insights into the activation mechanism of the alternative pathway C3-convertase.
Mol.Cell, 14, 2004
|
|
1RS6
 
 | | Rat neuronal NOS heme domain with D-lysine-D-nitroarginine amide bound | | Descriptor: | 5,6,7,8-TETRAHYDROBIOPTERIN, ACETATE ION, D-MANNITOL, ... | | Authors: | Flinspach, M, Li, H, Jamal, J, Yang, W, Huang, H, Silverman, R.B, Poulos, T.L. | | Deposit date: | 2003-12-09 | | Release date: | 2004-05-18 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Structures of the Neuronal and Endothelial Nitric Oxide Synthase Heme Domain with d-Nitroarginine-Containing Dipeptide Inhibitors Bound.
Biochemistry, 43, 2004
|
|
1RS7
 
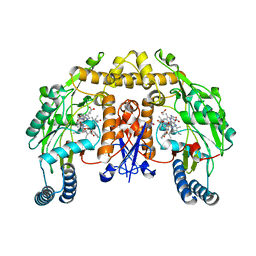 | | Rat neuronal NOS heme domain with D-phenylalanine-D-nitroarginine amide bound | | Descriptor: | 5,6,7,8-TETRAHYDROBIOPTERIN, ACETATE ION, D-MANNITOL, ... | | Authors: | Flinspach, M, Li, H, Jamal, J, Yang, W, Huang, H, Silverman, R.B, Poulos, T.L. | | Deposit date: | 2003-12-09 | | Release date: | 2004-05-18 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Structures of the Neuronal and Endothelial Nitric Oxide Synthase Heme Domain with d-Nitroarginine-Containing Dipeptide Inhibitors Bound.
Biochemistry, 43, 2004
|
|
1RS8
 
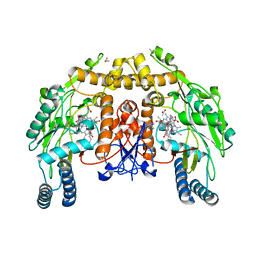 | | Bovine endothelial NOS heme domain with D-lysine-D-nitroarginine amide bound | | Descriptor: | 5,6,7,8-TETRAHYDROBIOPTERIN, ACETATE ION, CACODYLATE ION, ... | | Authors: | Flinspach, M, Li, H, Jamal, J, Yang, W, Huang, H, Silverman, R.B, Poulos, T.L. | | Deposit date: | 2003-12-09 | | Release date: | 2004-06-15 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structures of the neuronal and endothelial nitric oxide synthase heme domain with D-nitroarginine-containing dipeptide inhibitors bound.
Biochemistry, 43, 2004
|
|
1RS9
 
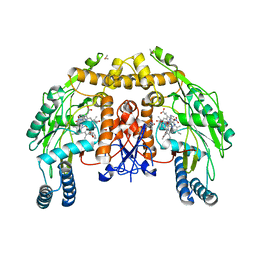 | | Bovine endothelial NOS heme domain with D-phenylalanine-D-nitroarginine amide bound | | Descriptor: | 5,6,7,8-TETRAHYDROBIOPTERIN, ACETATE ION, CACODYLATE ION, ... | | Authors: | Flinspach, M, Li, H, Jamal, J, Yang, W, Huang, H, Silverman, R.B, Poulos, T.L. | | Deposit date: | 2003-12-09 | | Release date: | 2004-06-15 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.22 Å) | | Cite: | Structures of the neuronal and endothelial nitric oxide synthase heme domain with D-nitroarginine-containing dipeptide inhibitors bound.
Biochemistry, 43, 2004
|
|
1RSB
 
 | |
1RSG
 
 | |
1RSM
 
 | | THE 2-ANGSTROMS RESOLUTION STRUCTURE OF A THERMOSTABLE RIBONUCLEASE A CHEMICALLY CROSS-LINKED BETWEEN LYSINE RESIDUES 7 AND 41 | | Descriptor: | DINITROPHENYLENE, RIBONUCLEASE A | | Authors: | Weber, P.C, Sheriff, S, Ohlendorf, D.H, Finzel, B.C, Salemme, F.R. | | Deposit date: | 1985-08-27 | | Release date: | 1986-01-21 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | The 2-A resolution structure of a thermostable ribonuclease A chemically cross-linked between lysine residues 7 and 41.
Proc.Natl.Acad.Sci.USA, 82, 1985
|
|
1RSN
 
 | | RIBONUCLEASE (RNASE SA) (E.C.3.1.4.8) COMPLEXED WITH EXO-2',3'-CYCLOPHOSPHOROTHIOATE | | Descriptor: | GUANOSINE-2',3'-CYCLOPHOSPHOROTHIOATE, RIBONUCLEASE SA, SULFATE ION | | Authors: | Sevcik, J, Dauter, Z, Lamzin, V.S, Wilson, K.S. | | Deposit date: | 1995-09-01 | | Release date: | 1995-12-07 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Complex of ribonuclease Sa with a cyclic nucleotide and a proposed model for the reaction intermediate.
Eur.J.Biochem., 216, 1993
|
|
1RSR
 
 | | azide complex of the diferrous F208A mutant R2 subunit of ribonucleotide reductase | | Descriptor: | AZIDE ION, FE (II) ION, MERCURY (II) ION, ... | | Authors: | Andersson, M.E, Hogbom, M, Rinaldo-Matthis, A, Andersson, K.K, Sjoberg, B.M, Nordlund, P. | | Deposit date: | 2003-12-10 | | Release date: | 2003-12-23 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | The Crystal Structure of an Azide Complex of the Diferrous R2 Subunit of Ribonucleotide Reductase Displays a Novel Carboxylate Shift with Important Mechanistic Implications for Diiron-Catalyzed Oxygen Activation
J.Am.Chem.Soc., 121, 1999
|
|
1RSS
 
 | |
