4YKO
 
 | |
4YKP
 
 | |
4YKQ
 
 | | Heat Shock Protein 90 Bound to CS301 | | Descriptor: | 1-(5-ethyl-2,4-dihydroxyphenyl)-1,3-dihydro-2H-benzimidazol-2-one, Heat shock protein HSP 90-alpha | | Authors: | Kang, Y.N, Stuckey, J.A. | | Deposit date: | 2015-03-04 | | Release date: | 2016-03-09 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.91 Å) | | Cite: | Structure of Heat Shock Protein 90 Bound to CS301
To Be Published
|
|
4YKR
 
 | | Heat Shock Protein 90 Bound to CS302 | | Descriptor: | 1-(2,4-dihydroxy-5-propylphenyl)-1,3-dihydro-2H-benzimidazol-2-one, GLYCEROL, Heat shock protein HSP 90-alpha | | Authors: | Kang, Y.N, Stuckey, J.A. | | Deposit date: | 2015-03-04 | | Release date: | 2016-03-09 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.61 Å) | | Cite: | Structure of Heat Shock Protein 90 Bound to CS302
To Be Published
|
|
4YKT
 
 | | Heat Shock Protein 90 Bound to CS307 | | Descriptor: | 1-(5-chloro-2,4-dihydroxyphenyl)-5-({[dihydroxy(pyridin-3-yl)-lambda~4~-sulfanyl]amino}methyl)-1,3-dihydro-2H-benzimidazol-2-one, Heat shock protein HSP 90-alpha | | Authors: | Kang, Y.N, Stuckey, J.A. | | Deposit date: | 2015-03-04 | | Release date: | 2016-03-09 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Structure of Heat Shock Protein 90 Bound to CS307
To Be Published
|
|
4YKU
 
 | | Heat Shock Protein 90 Bound to CS311 | | Descriptor: | 6-(2-chlorophenyl)-1,3,5-triazine-2,4-diamine, Heat shock protein HSP 90-alpha | | Authors: | Kang, Y.N, Stuckey, J.A. | | Deposit date: | 2015-03-04 | | Release date: | 2016-03-09 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structure of Heat Shock Protein 90 Bound to CS311
To Be Published
|
|
4YKW
 
 | | Heat Shock Protein 90 Bound to CS312 | | Descriptor: | 4-(2-chloro-4-nitrophenyl)-6-methylpyrimidin-2-amine, Heat shock protein HSP 90-alpha | | Authors: | Kang, Y.N, Stuckey, J.A. | | Deposit date: | 2015-03-04 | | Release date: | 2016-03-09 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Structure of Heat Shock Protein 90 Bound to CS312
To Be Published
|
|
4YKX
 
 | | Heat Shock Protein 90 Bound to CS318 | | Descriptor: | (5-chloro-2-hydroxyphenyl)(4-hydroxyphenyl)methanone, GLYCEROL, Heat shock protein HSP 90-alpha | | Authors: | Kang, Y.N, Stuckey, J.A. | | Deposit date: | 2015-03-04 | | Release date: | 2016-03-09 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structure of Heat Shock Protein 90 Bound to CS318
To Be Published
|
|
4YKY
 
 | | Heat Shock Protein 90 Bound to CS319 | | Descriptor: | (2,4-dihydroxyphenyl)(4-hydroxyphenyl)methanone, GLYCEROL, Heat shock protein HSP 90-alpha | | Authors: | Kang, Y.N, Stuckey, J.A. | | Deposit date: | 2015-03-04 | | Release date: | 2016-03-09 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.78 Å) | | Cite: | Structure of Heat Shock Protein 90 Bound to CS319
To Be Published
|
|
4YKZ
 
 | | Heat Shock Protein 90 Bound to CS320 | | Descriptor: | (2,4-dihydroxyphenyl)(3-hydroxyphenyl)methanone, Heat shock protein HSP 90-alpha | | Authors: | Kang, Y.N, Stuckey, J.A. | | Deposit date: | 2015-03-04 | | Release date: | 2016-03-09 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Structure of Heat Shock Protein 90 Bound to CS320
To Be Published
|
|
4YL0
 
 | | Crystal Structures of mPGES-1 Inhibitor Complexes | | Descriptor: | 2-(9-chloro-1H-phenanthro[9,10-d]imidazol-2-yl)benzene-1,3-dicarbonitrile, DI(HYDROXYETHYL)ETHER, GLUTATHIONE, ... | | Authors: | Luz, J.G, Antonysamy, S, Kuklish, S.L, Fisher, M.J. | | Deposit date: | 2015-03-04 | | Release date: | 2015-06-10 | | Last modified: | 2025-04-02 | | Method: | X-RAY DIFFRACTION (1.52 Å) | | Cite: | Crystal Structures of mPGES-1 Inhibitor Complexes Form a Basis for the Rational Design of Potent Analgesic and Anti-Inflammatory Therapeutics.
J.Med.Chem., 58, 2015
|
|
4YL1
 
 | | Crystal Structures of mPGES-1 Inhibitor Complexes | | Descriptor: | 5-(4-tert-butylphenyl)-1-[4-(propan-2-yloxy)phenyl]-1H-indole-2-carboxylic acid, DI(HYDROXYETHYL)ETHER, GLUTATHIONE, ... | | Authors: | Luz, J.G, Antonysamy, S, Kuklish, S.L, Fisher, M.J. | | Deposit date: | 2015-03-04 | | Release date: | 2015-06-10 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.41 Å) | | Cite: | Crystal Structures of mPGES-1 Inhibitor Complexes Form a Basis for the Rational Design of Potent Analgesic and Anti-Inflammatory Therapeutics.
J.Med.Chem., 58, 2015
|
|
4YL2
 
 | | Aerococcus viridans L-lactate oxidase Y191F mutant | | Descriptor: | FLAVIN MONONUCLEOTIDE, Lactate oxidase, PYRUVIC ACID | | Authors: | Rainer, D, Nidetzky, B, Wilson, D.K. | | Deposit date: | 2015-03-04 | | Release date: | 2015-08-26 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Speeding up the product release: a second-sphere contribution from Tyr191 to the reactivity of l-lactate oxidase revealed in crystallographic and kinetic studies of site-directed variants.
Febs J., 282, 2015
|
|
4YL3
 
 | | Crystal Structures of mPGES-1 Inhibitor Complexes | | Descriptor: | 5-[4-bromo-2-(2-chloro-6-fluorophenyl)-1H-imidazol-5-yl]-2-{[4-(trifluoromethyl)phenyl]ethynyl}pyridine, GLUTATHIONE, Prostaglandin E synthase, ... | | Authors: | Luz, J.G, Antonysamy, S, Kuklish, S.L, Fisher, M.J. | | Deposit date: | 2015-03-04 | | Release date: | 2015-07-08 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.41 Å) | | Cite: | Crystal Structures of mPGES-1 Inhibitor Complexes Form a Basis for the Rational Design of Potent Analgesic and Anti-Inflammatory Therapeutics.
J.Med.Chem., 58, 2015
|
|
4YL4
 
 | | 1.1 Angstrom resolution X-ray Crystallographic Structure of Psudoazurin | | Descriptor: | COPPER (II) ION, GLYCEROL, Pseudoazurin | | Authors: | Yamaguchi, T, Asamura, S, Takashina, A, Unno, M, Kohzuma, T. | | Deposit date: | 2015-03-05 | | Release date: | 2016-03-09 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.1 Å) | | Cite: | X-ray crystallographic evidence for the simultaneous presence of axial and rhombic sites in cupredoxins: atomic resolution X-ray crystal structure analysis of pseudoazurin and DFT modelling
Rsc Adv, 6, 2016
|
|
4YL5
 
 | |
4YL6
 
 | |
4YL7
 
 | |
4YL8
 
 | | Crystal structure of the Crumbs/Moesin complex | | Descriptor: | GLYCEROL, IODIDE ION, Moesin, ... | | Authors: | Wei, Z, Li, Y, Zhang, M. | | Deposit date: | 2015-03-05 | | Release date: | 2015-04-01 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Structural Basis for the Phosphorylation-regulated Interaction between the Cytoplasmic Tail of Cell Polarity Protein Crumbs and the Actin-binding Protein Moesin
J.Biol.Chem., 290, 2015
|
|
4YL9
 
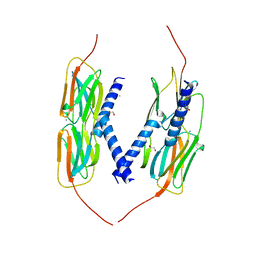 | | Crystal Structure of wild-type of hsp14.1 from Sulfolobus solfatataricus P2 | | Descriptor: | 1,2-ETHANEDIOL, CALCIUM ION, Heat shock protein Hsp20 | | Authors: | Liu, L, Chen, J.Y, Yun, C.H. | | Deposit date: | 2015-03-05 | | Release date: | 2015-11-04 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.353 Å) | | Cite: | Active-State Structures of a Small Heat-Shock Protein Revealed a Molecular Switch for Chaperone Function
Structure, 23, 2015
|
|
4YLA
 
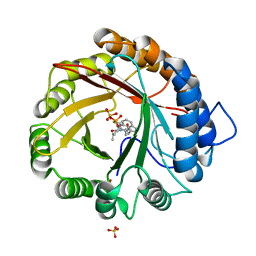 | | Crystal structure of the indole prenyltransferase MpnD complexed with indolactam V and DMSPP | | Descriptor: | (2S,5S)-5-(hydroxymethyl)-1-methyl-2-(propan-2-yl)-1,2,4,5,6,8-hexahydro-3H-[1,4]diazonino[7,6,5-cd]indol-3-one, Aromatic prenyltransferase, DIMETHYLALLYL S-THIOLODIPHOSPHATE, ... | | Authors: | Mori, T, Morita, H, Abe, I. | | Deposit date: | 2015-03-05 | | Release date: | 2016-03-16 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Manipulation of prenylation reactions by structure-based engineering of bacterial indolactam prenyltransferases.
Nat Commun, 7, 2016
|
|
4YLB
 
 | |
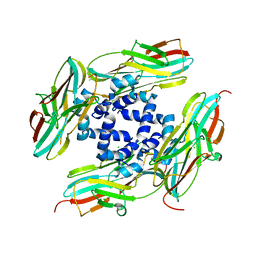
4YLC
 
 | |
4YLD
 
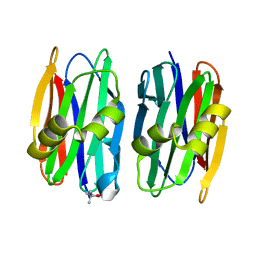 | | The crystal structure of Sclerotium Rolfsii lectin variant 1 (SSR1) | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, Sclerotium Rolfsii lectin variant 1 (SSR1) | | Authors: | Kantsadi, A.L, Peppa, V.I, Leonidas, D.D. | | Deposit date: | 2015-03-05 | | Release date: | 2015-07-01 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Molecular Cloning, Carbohydrate Specificity and the Crystal Structure of Two Sclerotium rolfsii Lectin Variants.
Molecules, 20, 2015
|
|
4YLE
 
 | | Crystal structure of an ABC transpoter solute binding protein (IPR025997) from Burkholderia multivorans (Bmul_1631, Target EFI-511115) with an unknown ligand modelled as alpha-D-erythrofuranose | | Descriptor: | Periplasmic binding protein/LacI transcriptional regulator, UNKNOWN LIGAND | | Authors: | Vetting, M.W, Al Obaidi, N.F, Toro, R, Morisco, L.L, Benach, J, Koss, J, Wasserman, S.R, Attonito, J.D, Scott Glenn, A, Chamala, S, Chowdhury, S, Lafleur, J, Love, J, Seidel, R.D, Whalen, K.L, Gerlt, J.A, Almo, S.C, Enzyme Function Initiative (EFI) | | Deposit date: | 2015-03-05 | | Release date: | 2015-04-15 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Crystal structure of an ABC transporter solute binding protein (IPR025997) from Burkholderia multivorans (Bmul_1631, Target EFI-511115) with an unknown ligand modelled as alpha-D-erythrofuranose
To be published
|
|
