4YGR
 
 | | Crystal structure of HAD phosphatase from Thermococcus onnurineus | | Descriptor: | 2-[N-CYCLOHEXYLAMINO]ETHANE SULFONIC ACID, Hydrolase, MAGNESIUM ION | | Authors: | Ngo, T.D, Le, B.V, Subramani, V.K, Nguyen, C.M.T, Lee, H.S, Cho, Y, Kim, K.K, Hwang, H.Y. | | Deposit date: | 2015-02-26 | | Release date: | 2015-04-22 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.703 Å) | | Cite: | Structural basis for the substrate selectivity of a HAD phosphatase from Thermococcus onnurineus NA1
Biochem.Biophys.Res.Commun., 461, 2015
|
|
4YGS
 
 | | Crystal structure of HAD phosphatase from Thermococcus onnurineus | | Descriptor: | CITRIC ACID, Hydrolase, MAGNESIUM ION | | Authors: | Ngo, T.D, Le, B.V, Subramani, V.K, Nguyen, C.M.T, Lee, H.S, Cho, Y, Kim, K.K, Hwang, H.Y. | | Deposit date: | 2015-02-26 | | Release date: | 2015-04-22 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structural basis for the substrate selectivity of a HAD phosphatase from Thermococcus onnurineus NA1
Biochem.Biophys.Res.Commun., 461, 2015
|
|
4YGT
 
 | |
4YGU
 
 | |
4YGV
 
 | | Reversal Agent for Dabigatran | | Descriptor: | GLYCEROL, aDabi-Fab2a heavy chain, aDabi-Fab2a light chain | | Authors: | Schiele, F, Nar, H. | | Deposit date: | 2015-02-26 | | Release date: | 2015-06-17 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (1.76 Å) | | Cite: | Structure-guided residence time optimization of a dabigatran reversal agent.
Mabs, 7, 2015
|
|
4YGW
 
 | | RNase S in complex with stabilized S peptide | | Descriptor: | 1-hydroxypropan-2-one, Ribonuclease A C2, S-peptide: ACE-LYS-GLU-THR-ALA-ALA-HCS-LYS-PHE-GLU-HCS-GLN-HIS-MET-ASP-SER, ... | | Authors: | Assem, N, Ferreira, D, Wolan, D.W, Dawson, P.E. | | Deposit date: | 2015-02-26 | | Release date: | 2015-07-01 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.18 Å) | | Cite: | Acetone-Linked Peptides: A Convergent Approach for Peptide Macrocyclization and Labeling.
Angew.Chem.Int.Ed.Engl., 54, 2015
|
|
4YGX
 
 | |
4YGY
 
 | |
4YGZ
 
 | |
4YH0
 
 | |
4YH1
 
 | |
4YH2
 
 | |
4YH3
 
 | | Crystal structure of human BRD4(1) in complex with 4-[(2E)-3-(4-methoxyphenyl)-2-phenylprop-2-enoyl]-3,4-dihydroquinoxalin-2(1H)-one (compound 19a) | | Descriptor: | 4-[(2E)-3-(4-methoxyphenyl)-2-phenylprop-2-enoyl]-3,4-dihydroquinoxalin-2(1H)-one, Bromodomain-containing protein 4 | | Authors: | White, A, Lakshminarasimhan, D, Suto, R.K. | | Deposit date: | 2015-02-26 | | Release date: | 2016-01-13 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Discovery of a new chemical series of BRD4(1) inhibitors using protein-ligand docking and structure-guided design.
Bioorg.Med.Chem.Lett., 25, 2015
|
|
4YH4
 
 | | Crystal structure of human BRD4(1) in complex with 4-[(5-phenylpyridin-3-yl)carbonyl]-3,4-dihydroquinoxalin-2(1H)-one (compound 19d) | | Descriptor: | 4-[(5-phenylpyridin-3-yl)carbonyl]-3,4-dihydroquinoxalin-2(1H)-one, Bromodomain-containing protein 4, GLYCEROL, ... | | Authors: | Lakshminarasimhan, D, White, A, Suto, R.K. | | Deposit date: | 2015-02-26 | | Release date: | 2016-01-13 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.33 Å) | | Cite: | Discovery of a new chemical series of BRD4(1) inhibitors using protein-ligand docking and structure-guided design.
Bioorg.Med.Chem.Lett., 25, 2015
|
|
4YH5
 
 | | Lipomyces starkeyi levoglucosan kinase bound to ADP and Manganese | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, ADENOSINE-5'-DIPHOSPHATE, Levoglucosan kinase, ... | | Authors: | Bacik, J.P. | | Deposit date: | 2015-02-26 | | Release date: | 2015-09-16 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Producing Glucose 6-Phosphate from Cellulosic Biomass: STRUCTURAL INSIGHTS INTO LEVOGLUCOSAN BIOCONVERSION.
J.Biol.Chem., 290, 2015
|
|
4YH6
 
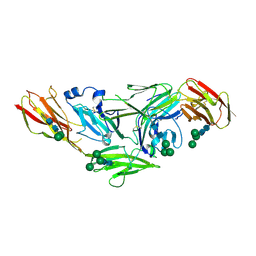 | | Crystal structure of IL1RAPL1 ectodomain | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Interleukin-1 receptor accessory protein-like 1, alpha-D-mannopyranose-(1-3)-[alpha-D-mannopyranose-(1-6)]beta-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Yamagata, A, Fukai, S. | | Deposit date: | 2015-02-27 | | Release date: | 2015-05-06 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Mechanisms of splicing-dependent trans-synaptic adhesion by PTP delta-IL1RAPL1/IL-1RAcP for synaptic differentiation.
Nat Commun, 6, 2015
|
|
4YH7
 
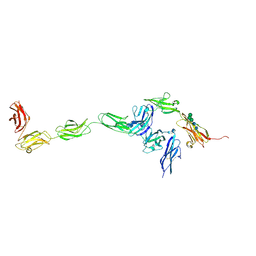 | | Crystal structure of PTPdelta ectodomain in complex with IL1RAPL1 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Interleukin-1 receptor accessory protein-like 1, ... | | Authors: | Yamagata, A, Fukai, S. | | Deposit date: | 2015-02-27 | | Release date: | 2015-05-06 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (4.4 Å) | | Cite: | Mechanisms of splicing-dependent trans-synaptic adhesion by PTP delta-IL1RAPL1/IL-1RAcP for synaptic differentiation.
Nat Commun, 6, 2015
|
|
4YH8
 
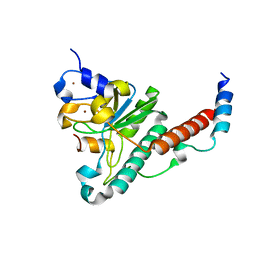 | | Structure of yeast U2AF complex | | Descriptor: | Splicing factor U2AF 23 kDa subunit, Splicing factor U2AF 59 kDa subunit, ZINC ION | | Authors: | Yoshida, H, Park, S.Y, Urano, T, Obayashi, E. | | Deposit date: | 2015-02-27 | | Release date: | 2015-08-12 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | A novel 3' splice site recognition by the two zinc fingers in the U2AF small subunit.
Genes Dev., 29, 2015
|
|
4YHA
 
 | |
4YHB
 
 | | Crystal structure of a siderophore utilization protein from T. fusca | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, FLAVIN-ADENINE DINUCLEOTIDE, Iron-chelator utilization protein, ... | | Authors: | Li, K, Bruner, S.D. | | Deposit date: | 2015-02-27 | | Release date: | 2015-07-15 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.8892 Å) | | Cite: | Structure and Mechanism of the Siderophore-Interacting Protein from the Fuscachelin Gene Cluster of Thermobifida fusca.
Biochemistry, 54, 2015
|
|
4YHC
 
 | | Crystal structure of the WD40 domain of SCAP from fission yeast | | Descriptor: | CITRIC ACID, Sterol regulatory element-binding protein cleavage-activating protein | | Authors: | Gong, X, Li, J.X, Wu, J.P, Yan, C.Y, Yan, N. | | Deposit date: | 2015-02-27 | | Release date: | 2015-04-01 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Structure of the WD40 domain of SCAP from fission yeast reveals the molecular basis for SREBP recognition.
Cell Res., 25, 2015
|
|
4YHD
 
 | | Staphylococcal alpha-hemolysin H35A mutant monomer | | Descriptor: | Alpha-hemolysin, CHLORIDE ION | | Authors: | Sugawara, T, Kato, K, Tanaka, Y, Yao, M. | | Deposit date: | 2015-02-27 | | Release date: | 2015-10-21 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.801 Å) | | Cite: | Structural basis for pore-forming mechanism of staphylococcal alpha-hemolysin
Toxicon, 108, 2015
|
|
4YHE
 
 | | NATIVE BACTEROIDETES-AFFILIATED GH5 CELLULASE LINKED WITH A POLYSACCHARIDE UTILIZATION LOCUS | | Descriptor: | GH5 | | Authors: | Naas, A.E, MacKenzie, A.K, Dalhus, B, Eijsink, V.G.H, Pope, P.B. | | Deposit date: | 2015-02-27 | | Release date: | 2015-05-20 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Structural Features of a Bacteroidetes-Affiliated Cellulase Linked with a Polysaccharide Utilization Locus.
Sci Rep, 5, 2015
|
|
4YHF
 
 | | Bruton's tyrosine kinase in complex with a t-butyl cyanoacrylamide inhibitor | | Descriptor: | (2S)-2-({(3R)-3-[4-amino-3-(4-phenoxyphenyl)-1H-pyrazolo[3,4-d]pyrimidin-1-yl]piperidin-1-yl}carbonyl)-4,4-dimethylpentanenitrile, 1,2-ETHANEDIOL, CHLORIDE ION, ... | | Authors: | Paavilainen, V.O, McFarland, J.M, Taunton, J. | | Deposit date: | 2015-02-27 | | Release date: | 2015-05-13 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Prolonged and tunable residence time using reversible covalent kinase inhibitors.
Nat.Chem.Biol., 11, 2015
|
|
4YHG
 
 | | NATIVE BACTEROIDETES-AFFILIATED GH5 CELLULASE LINKED WITH A POLYSACCHARIDE UTILIZATION LOCUS | | Descriptor: | GH5, beta-D-glucopyranose-(1-4)-beta-D-glucopyranose-(1-4)-beta-D-glucopyranose | | Authors: | Naas, A.E, MacKenzie, A.K, Dalhus, B, Eijsink, V.G.H, Pope, P.B. | | Deposit date: | 2015-02-27 | | Release date: | 2015-07-15 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structural Features of a Bacteroidetes-Affiliated Cellulase Linked with a Polysaccharide Utilization Locus.
Sci Rep, 5, 2015
|
|
