1F1T
 
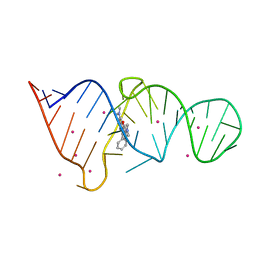 | | CRYSTAL STRUCTURE OF THE MALACHITE GREEN APTAMER COMPLEXED WITH TETRAMETHYL-ROSAMINE | | Descriptor: | MALACHITE GREEN APTAMER RNA, N,N'-TETRAMETHYL-ROSAMINE, STRONTIUM ION | | Authors: | Baugh, C, Grate, D, Wilson, C. | | Deposit date: | 2000-05-19 | | Release date: | 2000-09-04 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | 2.8 A crystal structure of the malachite green aptamer.
J.Mol.Biol., 301, 2000
|
|
1F1U
 
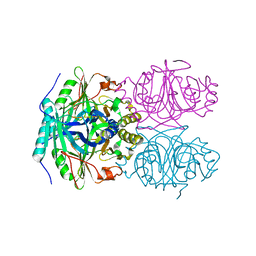 | | CRYSTAL STRUCTURE OF HOMOPROTOCATECHUATE 2,3-DIOXYGENASE FROM ARTHROBACTER GLOBIFORMIS (NATIVE, LOW TEMPERATURE) | | Descriptor: | HOMOPROTOCATECHUATE 2,3-DIOXYGENASE, MANGANESE (II) ION | | Authors: | Vetting, M.W, Lipscomb, J.D, Wackett, L.P, Que Jr, L, Ohlendorf, D.H. | | Deposit date: | 2000-05-19 | | Release date: | 2003-06-10 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Crystallographic comparison of manganese- and iron-dependent homoprotocatechuate 2,3-dioxygenases.
J.Bacteriol., 186, 2004
|
|
1F1V
 
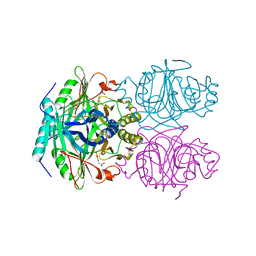 | | ANAEROBIC SUBSTRATE COMPLEX OF HOMOPROTOCATECHUATE 2,3-DIOXYGENASE FROM ARTHROBACTER GLOBIFORMIS. (COMPLEX WITH 3,4-DIHYDROXYPHENYLACETATE) | | Descriptor: | 2-(3,4-DIHYDROXYPHENYL)ACETIC ACID, HOMOPROTOCATECHUATE 2,3-DIOXYGENASE, MANGANESE (II) ION | | Authors: | Vetting, M.W, Lipscomb, J.D, Wackett, L.P, Que Jr, L, Ohlendorf, D.H. | | Deposit date: | 2000-05-20 | | Release date: | 2003-06-10 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystallographic comparison of manganese- and iron-dependent homoprotocatechuate 2,3-dioxygenases.
J.Bacteriol., 186, 2004
|
|
1F1W
 
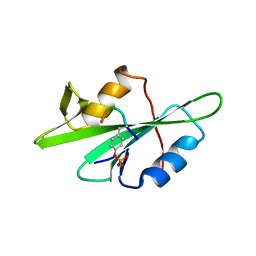 | | SRC SH2 THREF1TRP MUTANT COMPLEXED WITH THE PHOSPHOPEPTIDE S(PTR)VNVQN | | Descriptor: | PROTO-ONCOGENE TYROSINE-PROTEIN KINASE SRC, S(PTR)VNVQN PHOSPHOPEPTIDE | | Authors: | Kimber, M.S, Nachman, J, Cunningham, A.M, Gish, G.D, Pawson, T, Pai, E.F. | | Deposit date: | 2000-05-20 | | Release date: | 2000-07-06 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structural basis for specificity switching of the Src SH2 domain.
Mol.Cell, 5, 2000
|
|
1F1X
 
 | | CRYSTAL STRUCTURE OF HOMOPROTOCATECHUATE 2,3-DIOXYGENASE FROM BREVIBACTERIUM FUSCUM | | Descriptor: | HOMOPROTOCATECHUATE 2,3-DIOXYGENASE, HYDRATED FE | | Authors: | Vetting, M.W, Lipscomb, J.D, Wackett, L.P, Que Jr, L, Ohlendorf, D.H. | | Deposit date: | 2000-05-20 | | Release date: | 2003-06-10 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Crystallographic comparison of manganese- and iron-dependent homoprotocatechuate 2,3-dioxygenases.
J.Bacteriol., 186, 2004
|
|
1F20
 
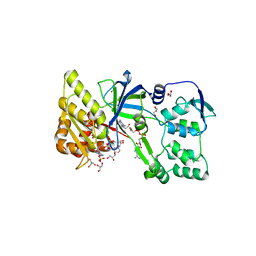 | | CRYSTAL STRUCTURE OF RAT NEURONAL NITRIC-OXIDE SYNTHASE FAD/NADP+ DOMAIN AT 1.9A RESOLUTION. | | Descriptor: | FLAVIN-ADENINE DINUCLEOTIDE, FORMIC ACID, GLYCEROL, ... | | Authors: | Zhang, J, Martasek, P, Masters, B.S, Kim, J.P. | | Deposit date: | 2000-05-22 | | Release date: | 2001-10-10 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal structure of the FAD/NADPH-binding domain of rat neuronal nitric-oxide synthase. Comparisons with NADPH-cytochrome P450 oxidoreductase.
J.Biol.Chem., 276, 2001
|
|
1F2E
 
 | | STRUCTURE OF SPHINGOMONAD, GLUTATHIONE S-TRANSFERASE COMPLEXED WITH GLUTATHIONE | | Descriptor: | GLUTATHIONE, GLUTATHIONE S-TRANSFERASE | | Authors: | Nishio, T, Watanabe, T, Patel, A, Wang, Y, Lau, P.C.K, Grochulski, P, Li, Y, Cygler, M. | | Deposit date: | 2000-05-24 | | Release date: | 2000-06-21 | | Last modified: | 2011-12-21 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Properties of a Sphingomonad and Marine Bacterium Beta-Class Glutathione S-Transferases and Crystal Structure of the Former Complex with Glutathione
To be published
|
|
1F2F
 
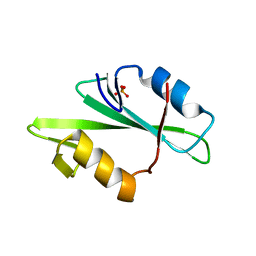 | | SRC SH2 THREF1TRP MUTANT | | Descriptor: | PHOSPHATE ION, PROTO-ONCOGENE TYROSINE-PROTEIN KINASE SRC | | Authors: | Kimber, M.S, Nachman, J, Cunningham, A.M, Gish, G.D, Pawson, T, Pai, E.F. | | Deposit date: | 2000-05-24 | | Release date: | 2000-07-06 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structural basis for specificity switching of the Src SH2 domain.
Mol.Cell, 5, 2000
|
|
1F2L
 
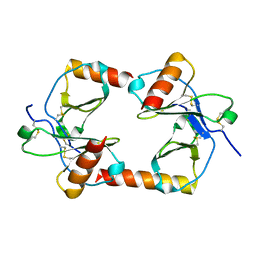 | |
1F2N
 
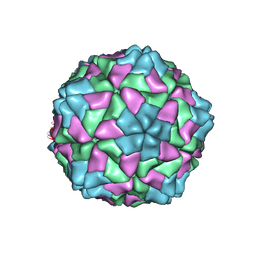 | | RICE YELLOW MOTTLE VIRUS | | Descriptor: | CALCIUM ION, CAPSID PROTEIN | | Authors: | Qu, C, Liljas, L, Opalka, N, Brugidou, C, Yeager, M, Beachy, R.N, Fauquet, C.M, Johnson, J.E, Lin, T. | | Deposit date: | 2000-05-26 | | Release date: | 2000-11-01 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | 3D domain swapping modulates the stability of members of an icosahedral virus group.
Structure Fold.Des., 8, 2000
|
|
1F2S
 
 | | CRYSTAL STRUCTURE OF THE COMPLEX FORMED BETWEEN BOVINE BETA-TRYPSIN AND MCTI-A, A TRYPSIN INHIBITOR OF SQUASH FAMILY AT 1.8 A RESOLUTION | | Descriptor: | CALCIUM ION, TRYPSIN, TRYPSIN INHIBITOR A | | Authors: | Zhu, Y, Huang, Q, Qian, M, Jia, Y, Tang, Y. | | Deposit date: | 2000-05-29 | | Release date: | 2000-06-05 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.79 Å) | | Cite: | Crystal structure of the complex formed between bovine beta-trypsin and MCTI-A, a trypsin inhibitor of squash family, at 1.8-A resolution.
J.Protein Chem., 18, 1999
|
|
1F2V
 
 | | CRYSTAL STRUCTURE ANALYSIS OF PRECORRIN-8X METHYLMUTASE OF AEROBIC VITAMIN B12 SYNTHESIS | | Descriptor: | PRECORRIN-8X METHYLMUTASE | | Authors: | Shipman, L.W, Li, D, Roessner, C.A, Scott, A.I, Sacchettini, J.C. | | Deposit date: | 2000-05-29 | | Release date: | 2001-07-18 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Crystal structure of precorrin-8x methyl mutase.
Structure, 9, 2001
|
|
1F2W
 
 | | THE MECHANISM OF CYANAMIDE HYDRATION CATALYZED BY CARBONIC ANHYDRASE II REVEALED BY CRYOGENIC X-RAY DIFFRACTION | | Descriptor: | 4-(HYDROXYMERCURY)BENZOIC ACID, CARBONIC ANHYDRASE II, CYANAMIDE, ... | | Authors: | Guerri, A, Briganti, F, Scozzafava, A, Supuran, C.T, Mangani, S. | | Deposit date: | 2000-05-30 | | Release date: | 2000-06-08 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Mechanism of cyanamide hydration catalyzed by carbonic anhydrase II suggested by cryogenic X-ray diffraction.
Biochemistry, 39, 2000
|
|
1F2X
 
 | |
1F30
 
 | | THE STRUCTURAL BASIS FOR DNA PROTECTION BY E. COLI DPS PROTEIN | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, DNA PROTECTION DURING STARVATION PROTEIN, ZINC ION | | Authors: | Luo, J, Liu, D, White, M.A, Fox, R.O. | | Deposit date: | 2000-05-31 | | Release date: | 2003-06-17 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.85 Å) | | Cite: | The Structural Basis for DNA Protection by E. coli Dps Protein
To be Published
|
|
1F32
 
 | | CRYSTAL STRUCTURE OF ASCARIS PEPSIN INHIBITOR-3 | | Descriptor: | MAJOR PEPSIN INHIBITOR PI-3 | | Authors: | Ng, K.K, Petersen, J.F, Cherney, M.M, Garen, C, James, M.N. | | Deposit date: | 2000-05-31 | | Release date: | 2001-02-01 | | Last modified: | 2018-04-04 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Structural basis for the inhibition of porcine pepsin by Ascaris pepsin inhibitor-3.
Nat.Struct.Biol., 7, 2000
|
|
1F33
 
 | |
1F34
 
 | | CRYSTAL STRUCTURE OF ASCARIS PEPSIN INHIBITOR-3 BOUND TO PORCINE PEPSIN | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, MAJOR PEPSIN INHIBITOR PI-3, PEPSIN A | | Authors: | Ng, K.K, Petersen, J.F, Cherney, M.M, Garen, C, James, M.N. | | Deposit date: | 2000-05-31 | | Release date: | 2001-02-01 | | Last modified: | 2018-04-04 | | Method: | X-RAY DIFFRACTION (2.45 Å) | | Cite: | Structural basis for the inhibition of porcine pepsin by Ascaris pepsin inhibitor-3.
Nat.Struct.Biol., 7, 2000
|
|
1F35
 
 | |
1F36
 
 | |
1F38
 
 | |
1F3A
 
 | | CRYSTAL STRUCTURE OF MGSTA1-1 IN COMPLEX WITH GSH | | Descriptor: | GLUTATHIONE, GLUTATHIONE S-TRANSFERASE YA CHAIN | | Authors: | Gu, Y, Singh, S.V, Ji, X. | | Deposit date: | 2000-06-01 | | Release date: | 2000-10-18 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Residue R216 and catalytic efficiency of a murine class alpha glutathione S-transferase toward benzo[a]pyrene 7(R),8(S)-diol 9(S), 10(R)-epoxide.
Biochemistry, 39, 2000
|
|
1F3B
 
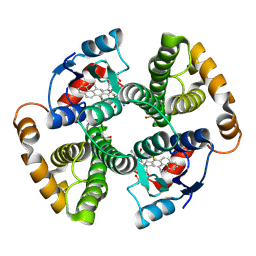 | | CRYSTAL STRUCTURE OF MGSTA1-1 IN COMPLEX WITH GLUTATHIONE CONJUGATE OF BENZO[A]PYRENE EPOXIDE | | Descriptor: | 2-AMINO-4-[1-(CARBOXYMETHYL-CARBAMOYL)-2-(9-HYDROXY-7,8-DIOXO-7,8,9,10-TETRAHYDRO-BENZO[DEF]CHRYSEN-10-YLSULFANYL)-ETHYLCARBAMOYL]-BUTYRIC ACID, GLUTATHIONE S-TRANSFERASE YA CHAIN | | Authors: | Gu, Y, Singh, S.V, Ji, X. | | Deposit date: | 2000-06-01 | | Release date: | 2000-10-18 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Residue R216 and catalytic efficiency of a murine class alpha glutathione S-transferase toward benzo[a]pyrene 7(R),8(S)-diol 9(S), 10(R)-epoxide.
Biochemistry, 39, 2000
|
|
1F3D
 
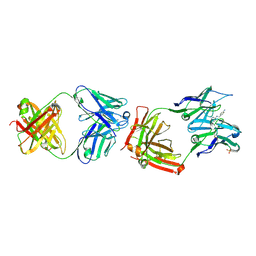 | | CATALYTIC ANTIBODY 4B2 IN COMPLEX WITH ITS AMIDINIUM HAPTEN. | | Descriptor: | 2-(4-AMINOBENZYLAMINO)-3,4,5,6-TETRAHYDROPYRIDINIUM, CATALYTIC ANTIBODY 4B2, SULFATE ION | | Authors: | Golinelli-Pimpaneau, B, Goncalves, O, Dintinger, T, Blanchard, D, Knossow, M, Tellier, C. | | Deposit date: | 2000-06-02 | | Release date: | 2000-09-13 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.87 Å) | | Cite: | Structural evidence for a programmed general base in the active site of a catalytic antibody.
Proc.Natl.Acad.Sci.USA, 97, 2000
|
|
1F3E
 
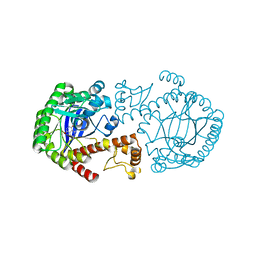 | | A NEW TARGET FOR SHIGELLOSIS: RATIONAL DESIGN AND CRYSTALLOGRAPHIC STUDIES OF INHIBITORS OF TRNA-GUANINE TRANSGLYCOSYLASE | | Descriptor: | 3,5-DIAMINOPHTHALHYDRAZIDE, QUEUINE TRNA-RIBOSYLTRANSFERASE, ZINC ION | | Authors: | Graedler, U, Gerber, H.-D, Goodenough-Lashua, D.M, Garcia, G.A.G, Ficner, R, Reuter, K, Stubbs, M.T, Klebe, G. | | Deposit date: | 2000-06-02 | | Release date: | 2000-06-15 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | A new target for shigellosis: rational design and crystallographic studies of inhibitors of tRNA-guanine transglycosylase.
J.Mol.Biol., 306, 2001
|
|
