1DJW
 
 | | PHOSPHOINOSITIDE-SPECIFIC PHOSPHOLIPASE C-DELTA1 FROM RAT COMPLEXED WITH INOSITOL-2-METHYLENE-1,2-CYCLIC-MONOPHOSPHONATE | | Descriptor: | ACETATE ION, CALCIUM ION, INOSITOL-2-METHYLENE-1,2-CYCLIC-MONOPHOSPHATE, ... | | Authors: | Essen, L.-O, Perisic, O, Williams, R.L. | | Deposit date: | 1996-08-24 | | Release date: | 1997-07-23 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.45 Å) | | Cite: | Structural mapping of the catalytic mechanism for a mammalian phosphoinositide-specific phospholipase C.
Biochemistry, 36, 1997
|
|
1DJX
 
 | | PHOSPHOINOSITIDE-SPECIFIC PHOSPHOLIPASE C-DELTA1 FROM RAT COMPLEXED WITH INOSITOL-1,4,5-TRISPHOSPHATE | | Descriptor: | ACETATE ION, CALCIUM ION, D-MYO-INOSITOL-1,4,5-TRIPHOSPHATE, ... | | Authors: | Essen, L.-O, Perisic, O, Williams, R.L. | | Deposit date: | 1996-08-24 | | Release date: | 1997-07-07 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structural mapping of the catalytic mechanism for a mammalian phosphoinositide-specific phospholipase C.
Biochemistry, 36, 1997
|
|
1DJY
 
 | | PHOSPHOINOSITIDE-SPECIFIC PHOSPHOLIPASE C-DELTA1 FROM RAT COMPLEXED WITH INOSITOL-2,4,5-TRISPHOSPHATE | | Descriptor: | ACETATE ION, CALCIUM ION, D-MYO-INOSITOL-2,4,5-TRIPHOSPHATE, ... | | Authors: | Essen, L.-O, Perisic, O, Williams, R.L. | | Deposit date: | 1996-08-24 | | Release date: | 1997-07-07 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structural mapping of the catalytic mechanism for a mammalian phosphoinositide-specific phospholipase C.
Biochemistry, 36, 1997
|
|
1DJZ
 
 | | PHOSPHOINOSITIDE-SPECIFIC PHOSPHOLIPASE C-DELTA1 FROM RAT COMPLEXED WITH INOSITOL-4,5-BISPHOSPHATE | | Descriptor: | ACETATE ION, CALCIUM ION, D-MYO-INOSITOL-4,5-BISPHOSPHATE, ... | | Authors: | Essen, L.-O, Perisic, O, Williams, R.L. | | Deposit date: | 1996-08-24 | | Release date: | 1997-07-07 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.95 Å) | | Cite: | Structural mapping of the catalytic mechanism for a mammalian phosphoinositide-specific phospholipase C.
Biochemistry, 36, 1997
|
|
1DK0
 
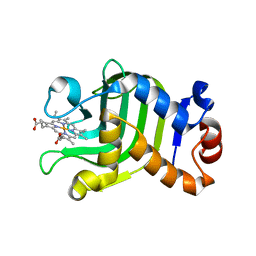 | | CRYSTAL STRUCTURE OF THE HEMOPHORE HASA FROM SERRATIA MARCESCENS CRYSTAL FORM P2(1), PH8 | | Descriptor: | HEME-BINDING PROTEIN A, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Arnoux, P, Haser, R, Izadi-Pruneyre, N, Lecroisey, A, Czjzek, M. | | Deposit date: | 1999-12-06 | | Release date: | 2000-12-27 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.77 Å) | | Cite: | Functional aspects of the heme bound hemophore HasA by structural analysis of various crystal forms.
Proteins, 41, 2000
|
|
1DK4
 
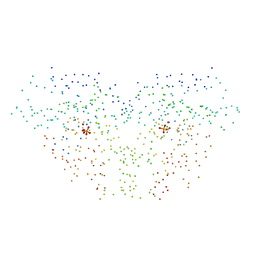 | | CRYSTAL STRUCTURE OF MJ0109 GENE PRODUCT INOSITOL MONOPHOSPHATASE | | Descriptor: | INOSITOL MONOPHOSPHATASE, PHOSPHATE ION, ZINC ION | | Authors: | Stec, B, Yang, H, Johnson, K.A, Chen, L, Roberts, M.F. | | Deposit date: | 1999-12-06 | | Release date: | 2000-11-08 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | MJ0109 is an enzyme that is both an inositol monophosphatase and the 'missing' archaeal fructose-1,6-bisphosphatase.
Nat.Struct.Biol., 7, 2000
|
|
1DK5
 
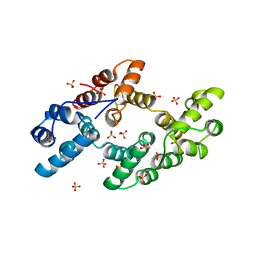 | | CRYSTAL STRUCTURE OF ANNEXIN 24(CA32) FROM CAPSICUM ANNUUM | | Descriptor: | ANNEXIN 24(CA32), SULFATE ION | | Authors: | Hofmann, A, Proust, J, Dorowski, A, Schantz, R, Huber, R. | | Deposit date: | 1999-12-06 | | Release date: | 2000-12-06 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Annexin 24 from Capsicum annuum. X-ray structure and biochemical characterization.
J.Biol.Chem., 275, 2000
|
|
1DK7
 
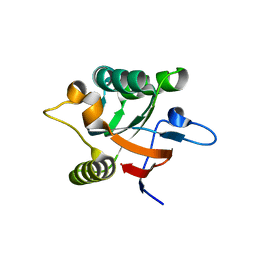 | |
1DK8
 
 | | CRYSTAL STRUCTURE OF THE RGS-HOMOLOGOUS DOMAIN OF AXIN | | Descriptor: | 2,3-DIHYDROXY-1,4-DITHIOBUTANE, AXIN, GLYCEROL, ... | | Authors: | Spink, K.E, Polakis, P, Weis, W.I. | | Deposit date: | 1999-12-06 | | Release date: | 2000-07-12 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.57 Å) | | Cite: | Structural basis of the Axin-adenomatous polyposis coli interaction.
EMBO J., 19, 2000
|
|
1DKD
 
 | |
1DKF
 
 | | CRYSTAL STRUCTURE OF A HETERODIMERIC COMPLEX OF RAR AND RXR LIGAND-BINDING DOMAINS | | Descriptor: | 4-[(4,4-DIMETHYL-1,2,3,4-TETRAHYDRO-[1,2']BINAPTHALENYL-7-CARBONYL)-AMINO]-BENZOIC ACID, OLEIC ACID, PROTEIN (RETINOIC ACID RECEPTOR-ALPHA), ... | | Authors: | Bourguet, W, Vivat, V, Wurtz, J.M, Chambon, P, Gronemeyer, H, Moras, D, Structural Proteomics in Europe (SPINE) | | Deposit date: | 1999-12-07 | | Release date: | 2000-04-19 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Crystal structure of a heterodimeric complex of RAR and RXR ligand-binding domains.
Mol.Cell, 5, 2000
|
|
1DKG
 
 | |
1DKH
 
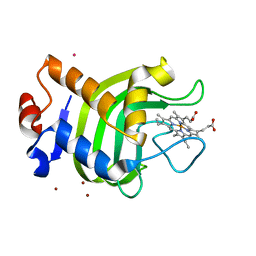 | | CRYSTAL STRUCTURE OF THE HEMOPHORE HASA, PH 6.5 | | Descriptor: | HEME-BINDING PROTEIN A, PROTOPORPHYRIN IX CONTAINING FE, SAMARIUM (III) ION, ... | | Authors: | Arnoux, P, Haser, R, Izadi-Pruneyre, N, Lecroisey, A, Czjzek, M. | | Deposit date: | 1999-12-07 | | Release date: | 2000-12-27 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Functional aspects of the heme bound hemophore HasA by structural analysis of various crystal forms.
Proteins, 41, 2000
|
|
1DKI
 
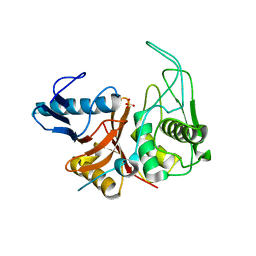 | | CRYSTAL STRUCTURE OF THE ZYMOGEN FORM OF STREPTOCOCCAL PYROGENIC EXOTOXIN B ACTIVE SITE (C47S) MUTANT | | Descriptor: | PYROGENIC EXOTOXIN B ZYMOGEN, SULFATE ION | | Authors: | Kagawa, T.F, Cooney, J.C, Baker, H.M, McSweeney, S, Liu, M, Gubba, S, Musser, J.M, Baker, E.N. | | Deposit date: | 1999-12-07 | | Release date: | 2000-03-01 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Crystal structure of the zymogen form of the group A Streptococcus virulence factor SpeB: an integrin-binding cysteine protease.
Proc.Natl.Acad.Sci.USA, 97, 2000
|
|
1DKJ
 
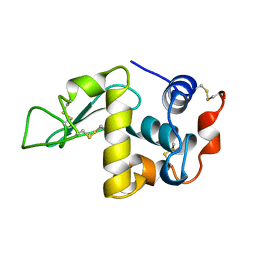 | | BOBWHITE QUAIL LYSOZYME | | Descriptor: | LYSOZYME | | Authors: | Jeffrey, P.D, Sheriff, S. | | Deposit date: | 1996-01-10 | | Release date: | 1996-07-11 | | Last modified: | 2024-06-05 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Refined structures of bobwhite quail lysozyme uncomplexed and complexed with the HyHEL-5 Fab fragment.
Proteins, 26, 1996
|
|
1DKK
 
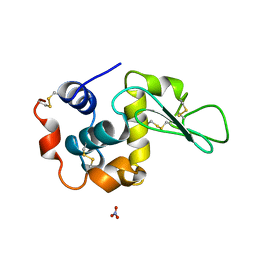 | | BOBWHITE QUAIL LYSOZYME WITH NITRATE | | Descriptor: | LYSOZYME, NITRATE ION | | Authors: | Jeffrey, P.D, Sheriff, S. | | Deposit date: | 1996-01-10 | | Release date: | 1996-07-11 | | Last modified: | 2018-04-18 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Refined structures of bobwhite quail lysozyme uncomplexed and complexed with the HyHEL-5 Fab fragment.
Proteins, 26, 1996
|
|
1DKL
 
 | |
1DKM
 
 | | CRYSTAL STRUCTURE OF ESCHERICHIA COLI PHYTASE AT PH 6.6 WITH HG2+ CATION ACTING AS AN INTERMOLECULAR BRIDGE | | Descriptor: | MERCURY (II) ION, PHYTASE | | Authors: | Lim, D, Golovan, S, Forsberg, C.W, Jia, Z. | | Deposit date: | 1999-12-08 | | Release date: | 2000-08-02 | | Last modified: | 2021-11-03 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Crystal structures of Escherichia coli phytase and its complex with phytate.
Nat.Struct.Biol., 7, 2000
|
|
1DKN
 
 | | CRYSTAL STRUCTURE OF ESCHERICHIA COLI PHYTASE AT PH 5.0 WITH HG2+ CATION ACTING AS AN INTERMOLECULAR BRIDGE | | Descriptor: | MERCURY (II) ION, PHYTASE | | Authors: | Lim, D, Golovan, S, Forsberg, C.W, Jia, Z. | | Deposit date: | 1999-12-08 | | Release date: | 2000-08-03 | | Last modified: | 2021-11-03 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Crystal structures of Escherichia coli phytase and its complex with phytate.
Nat.Struct.Biol., 7, 2000
|
|
1DKO
 
 | | CRYSTAL STRUCTURE OF TUNGSTATE COMPLEX OF ESCHERICHIA COLI PHYTASE AT PH 6.6 WITH TUNGSTATE BOUND AT THE ACTIVE SITE AND WITH HG2+ CATION ACTING AS AN INTERMOLECULAR BRIDGE | | Descriptor: | MERCURY (II) ION, PHYTASE, TUNGSTATE(VI)ION | | Authors: | Lim, D, Golovan, S, Forsberg, C.W, Jia, Z. | | Deposit date: | 1999-12-08 | | Release date: | 2000-08-03 | | Last modified: | 2021-11-03 | | Method: | X-RAY DIFFRACTION (2.38 Å) | | Cite: | Crystal structures of Escherichia coli phytase and its complex with phytate.
Nat.Struct.Biol., 7, 2000
|
|
1DKP
 
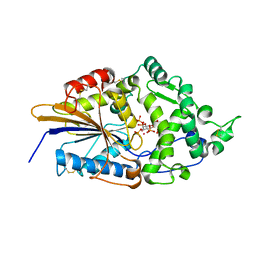 | | CRYSTAL STRUCTURE OF PHYTATE COMPLEX OF ESCHERICHIA COLI PHYTASE AT PH 6.6. PHYTATE IS BOUND WITH ITS 3-PHOSPHATE IN THE ACTIVE SITE. HG2+ CATION ACTS AS AN INTERMOLECULAR BRIDGE | | Descriptor: | INOSITOL HEXAKISPHOSPHATE, MERCURY (II) ION, PHYTASE | | Authors: | Lim, D, Golovan, S, Forsberg, C.W, Jia, Z. | | Deposit date: | 1999-12-08 | | Release date: | 2000-08-03 | | Last modified: | 2021-11-03 | | Method: | X-RAY DIFFRACTION (2.28 Å) | | Cite: | Crystal structures of Escherichia coli phytase and its complex with phytate.
Nat.Struct.Biol., 7, 2000
|
|
1DKQ
 
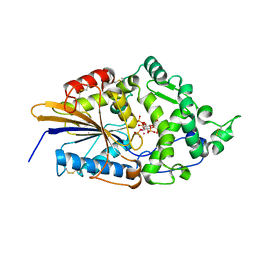 | | CRYSTAL STRUCTURE OF PHYTATE COMPLEX ESCHERICHIA COLI PHYTASE AT PH 5.0. PHYTATE IS BOUND WITH ITS 3-PHOSPHATE IN THE ACTIVE SITE. HG2+ CATION ACTS AS AN INTERMOLECULAR BRIDGE | | Descriptor: | INOSITOL HEXAKISPHOSPHATE, MERCURY (II) ION, PHYTASE | | Authors: | Lim, D, Golovan, S, Forsberg, C.W, Jia, Z. | | Deposit date: | 1999-12-08 | | Release date: | 2000-08-03 | | Last modified: | 2021-11-03 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Crystal structures of Escherichia coli phytase and its complex with phytate.
Nat.Struct.Biol., 7, 2000
|
|
1DKR
 
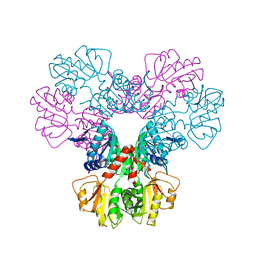 | | CRYSTAL STRUCTURES OF BACILLUS SUBTILIS PHOSPHORIBOSYLPYROPHOSPHATE SYNTHETASE: MOLECULAR BASIS OF ALLOSTERIC INHIBITION AND ACTIVATION. | | Descriptor: | PHOSPHORIBOSYL PYROPHOSPHATE SYNTHETASE, SULFATE ION | | Authors: | Eriksen, T.A, Kadziola, A, Bentsen, A.-K, Harlow, K.W, Larsen, S. | | Deposit date: | 1999-12-08 | | Release date: | 2000-04-05 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structural basis for the function of Bacillus subtilis phosphoribosyl-pyrophosphate synthetase.
Nat.Struct.Biol., 7, 2000
|
|
1DKU
 
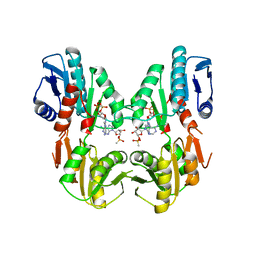 | | CRYSTAL STRUCTURES OF BACILLUS SUBTILIS PHOSPHORIBOSYLPYROPHOSPHATE SYNTHETASE: MOLECULAR BASIS OF ALLOSTERIC INHIBITION AND ACTIVATION. | | Descriptor: | METHYL PHOSPHONIC ACID ADENOSINE ESTER, PHOSPHOMETHYLPHOSPHONIC ACID ADENOSYL ESTER, PROTEIN (PHOSPHORIBOSYL PYROPHOSPHATE SYNTHETASE) | | Authors: | Eriksen, T.A, Kadziola, A, Bentsen, A.-K, Harlow, K.W, Larsen, S. | | Deposit date: | 1999-12-08 | | Release date: | 2000-04-05 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structural basis for the function of Bacillus subtilis phosphoribosyl-pyrophosphate synthetase.
Nat.Struct.Biol., 7, 2000
|
|
1DKW
 
 | | CRYSTAL STRUCTURE OF TRIOSE-PHOSPHATE ISOMERASE WITH MODIFIED SUBSTRATE BINDING SITE | | Descriptor: | TERTIARY-BUTYL ALCOHOL, TRIOSEPHOSPHATE ISOMERASE | | Authors: | Norledge, B.V, Lambeir, A.M, Abagyan, R.A, Rottman, A, Fernandez, A.M, Filimonov, V.V, Peter, M.G, Wierenga, R.K. | | Deposit date: | 1999-12-08 | | Release date: | 2000-11-03 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.65 Å) | | Cite: | Modeling, mutagenesis, and structural studies on the fully conserved phosphate-binding loop (loop 8) of triosephosphate isomerase: toward a new substrate specificity.
Proteins, 42, 2001
|
|
