1KXX
 
 | |
1KXY
 
 | |
1KXZ
 
 | | MT0146, the Precorrin-6y methyltransferase (CbiT) homolog from M. Thermoautotrophicum, P1 spacegroup | | Descriptor: | Precorrin-6y methyltransferase/putative decarboxylase | | Authors: | Keller, J.P, Smith, P.M, Benach, J, Christendat, D, DeTitta, G, Hunt, J.F. | | Deposit date: | 2002-02-01 | | Release date: | 2002-11-27 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | The Crystal Structure of MT0146/CbiT Suggests that the Putative
Precorrin-8W Decarboxylase is a Methyltransferase
Structure, 10, 2002
|
|
1KY0
 
 | | METHIONINE CORE MUTANT OF T4 LYSOZYME | | Descriptor: | 2-HYDROXYETHYL DISULFIDE, CHLORIDE ION, LYSOZYME | | Authors: | Gassner, N.C, Baase, W.A, Mooers, B.H, Busam, R.D, Weaver, L.H, Lindstrom, J.D, Quillin, M.L, Matthews, B.W. | | Deposit date: | 2002-02-01 | | Release date: | 2003-06-03 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.97 Å) | | Cite: | Multiple methionine substitutions are tolerated in T4 lysozyme and have coupled effects on folding and stability
BIOPHYS.CHEM., 100, 2003
|
|
1KY1
 
 | | METHIONINE CORE MUTANT OF T4 LYSOZYME | | Descriptor: | 2-HYDROXYETHYL DISULFIDE, CHLORIDE ION, LYSOZYME | | Authors: | Gassner, N.C, Baase, W.A, Mooers, B.H, Busam, R.D, Weaver, L.H, Lindstrom, J.D, Quillin, M.L, Matthews, B.W. | | Deposit date: | 2002-02-01 | | Release date: | 2003-06-03 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Multiple methionine substitutions are tolerated in T4 lysozyme and have coupled effects on folding and stability
BIOPHYS.CHEM., 100, 2003
|
|
1KY2
 
 | | GPPNHP-BOUND YPT7P AT 1.6 A RESOLUTION | | Descriptor: | GTP-BINDING PROTEIN YPT7P, MAGNESIUM ION, PHOSPHOAMINOPHOSPHONIC ACID-GUANYLATE ESTER | | Authors: | Constantinescu, A.-T, Rak, A, Scheidig, A.J. | | Deposit date: | 2002-02-02 | | Release date: | 2002-06-05 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Rab-subfamily-specific regions of Ypt7p are structurally different from other RabGTPases.
Structure, 10, 2002
|
|
1KY3
 
 | | GDP-BOUND YPT7P AT 1.35 A RESOLUTION | | Descriptor: | GTP-BINDING PROTEIN YPT7P, GUANOSINE-5'-DIPHOSPHATE, MAGNESIUM ION | | Authors: | Constantinescu, A.-T, Rak, A, Scheidig, A.J. | | Deposit date: | 2002-02-02 | | Release date: | 2002-06-05 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.35 Å) | | Cite: | Rab-subfamily-specific regions of Ypt7p are structurally different from other RabGTPases.
Structure, 10, 2002
|
|
1KY8
 
 | | Crystal Structure of the Non-phosphorylating glyceraldehyde-3-phosphate Dehydrogenase | | Descriptor: | NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, SODIUM ION, glyceraldehyde-3-phosphate dehydrogenase | | Authors: | Pohl, E, Brunner, N, Wilmanns, M, Hensel, R. | | Deposit date: | 2002-02-04 | | Release date: | 2003-02-04 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | The Crystal Structure of the Allosteric Non-phosphorylating
glyceraldehyde-3-phosphate Dehydrogenase from the Hyperthermophilic
Archaeum Thermoproteus tenax
J.Biol.Chem., 277, 2002
|
|
1KY9
 
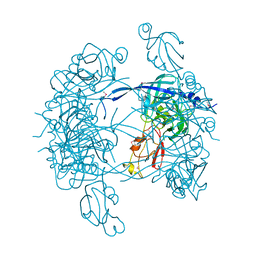 | | Crystal Structure of DegP (HtrA) | | Descriptor: | PROTEASE DO | | Authors: | Krojer, T, Garrido-Franco, M, Huber, R, Ehrmann, M, Clausen, T. | | Deposit date: | 2002-02-04 | | Release date: | 2002-04-03 | | Last modified: | 2021-10-27 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Crystal structure of DegP (HtrA) reveals a new protease-chaperone machine.
Nature, 416, 2002
|
|
1KYC
 
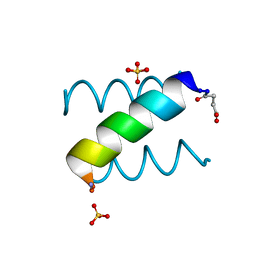 | |
1KYF
 
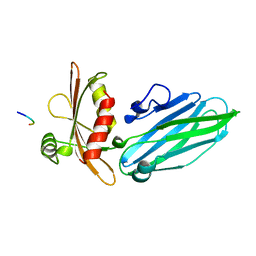 | |
1KYN
 
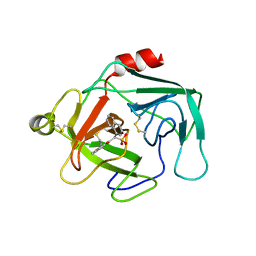 | | Cathepsin-G | | Descriptor: | (2-NAPHTHALEN-2-YL-1-NAPHTHALEN-1-YL-2-OXO-ETHYL)-PHOSPHONIC ACID, cathepsin G | | Authors: | Greco, M.N, Hawkins, M.J, Powell, E.T, Almond Jr, H.R, Corcoran, T.W, De Garavilla, L, Kauffman, J.A, Recacha, R, Chattopadhyay, D, Andrade-Gordon, P, Maryanoff, B.E. | | Deposit date: | 2002-02-05 | | Release date: | 2002-05-01 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (3.5 Å) | | Cite: | Nonpeptide inhibitors of cathepsin G: optimization of a novel beta-ketophosphonic acid lead by structure-based drug design.
J.Am.Chem.Soc., 124, 2002
|
|
1KYP
 
 | | Crystal Structure of an Apo Green Fluorescent Protein Zn Biosensor | | Descriptor: | Green Fluorescent Protein, MAGNESIUM ION | | Authors: | Barondeau, D.P, Kassmann, C.J, Tainer, J.A, Getzoff, E.D. | | Deposit date: | 2002-02-05 | | Release date: | 2002-04-10 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.35 Å) | | Cite: | Structural chemistry of a green fluorescent protein Zn biosensor.
J.Am.Chem.Soc., 124, 2002
|
|
1KYQ
 
 | | Met8p: A bifunctional NAD-dependent dehydrogenase and ferrochelatase involved in siroheme synthesis. | | Descriptor: | NICOTINAMIDE-ADENINE-DINUCLEOTIDE, Siroheme biosynthesis protein MET8 | | Authors: | Schubert, H.L, Raux, E, Brindley, A.A, Wilson, K.S, Hill, C.P, Warren, M.J. | | Deposit date: | 2002-02-05 | | Release date: | 2002-05-08 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | The structure of Saccharomyces cerevisiae Met8p, a bifunctional dehydrogenase and ferrochelatase.
EMBO J., 21, 2002
|
|
1KYR
 
 | | Crystal Structure of a Cu-bound Green Fluorescent Protein Zn Biosensor | | Descriptor: | COPPER (II) ION, Green Fluorescent Protein, MAGNESIUM ION | | Authors: | Barondeau, D.P, Kassmann, C.J, Tainer, J.A, Getzoff, E.D. | | Deposit date: | 2002-02-05 | | Release date: | 2002-04-10 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Structural chemistry of a green fluorescent protein Zn biosensor.
J.Am.Chem.Soc., 124, 2002
|
|
1KYS
 
 | | Crystal Structure of a Zn-bound Green Fluorescent Protein Biosensor | | Descriptor: | Green Fluorescent Protein, ZINC ION | | Authors: | Barondeau, D.P, Kassmann, C.J, Tainer, J.A, Getzoff, E.D. | | Deposit date: | 2002-02-05 | | Release date: | 2002-04-10 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.44 Å) | | Cite: | Structural chemistry of a green fluorescent protein Zn biosensor.
J.Am.Chem.Soc., 124, 2002
|
|
1KYT
 
 | | Crystal Structure of Thermoplasma acidophilum 0175 (APC014) | | Descriptor: | CALCIUM ION, hypothetical protein TA0175 | | Authors: | Kim, Y, Joachimiak, A, Edwards, A, Xu, X, Pennycooke, M, Gu, J, Cheung, F, Christendat, D, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2002-02-05 | | Release date: | 2003-01-21 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Crystal Structure of Thermoplasma acidophilum 0175 (APC014)
To be published
|
|
1KYW
 
 | | Crystal Structure Analysis of Caffeic Acid/5-hydroxyferulic acid 3/5-O-methyltransferase in complex with 5-hydroxyconiferaldehyde | | Descriptor: | 5-(3,3-DIHYDROXYPROPENY)-3-METHOXY-BENZENE-1,2-DIOL, Caffeic acid 3-O-methyltransferase, S-ADENOSYL-L-HOMOCYSTEINE | | Authors: | Zubieta, C, Kota, P, Ferrer, J.-L, Dixon, R.A, Noel, J.P. | | Deposit date: | 2002-02-06 | | Release date: | 2002-08-28 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structural basis for the modulation of lignin monomer methylation by caffeic acid/5-hydroxyferulic acid 3/5-O-methyltransferase.
Plant Cell, 14, 2002
|
|
1KYZ
 
 | | Crystal Structure Analysis of Caffeic acid/5-hydroxyferulic acid 3/5-O-methyltransferase Ferulic Acid Complex | | Descriptor: | 3-(4-HYDROXY-3-METHOXYPHENYL)-2-PROPENOIC ACID, Caffeic acid 3-O-methyltransferase, S-ADENOSYL-L-HOMOCYSTEINE | | Authors: | Zubieta, C, Kota, P, Ferrer, J.-L, Dixon, R.A, Noel, J.P. | | Deposit date: | 2002-02-06 | | Release date: | 2002-08-28 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structural basis for the modulation of lignin monomer methylation by caffeic acid/5-hydroxyferulic acid 3/5-O-methyltransferase.
Plant Cell, 14, 2002
|
|
1KZ7
 
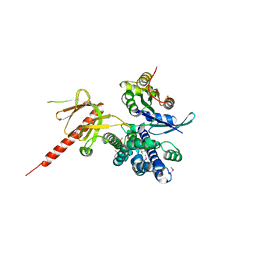 | | Crystal Structure of the DH/PH Fragment of Murine Dbs in Complex with the Placental Isoform of Human Cdc42 | | Descriptor: | CDC42 HOMOLOG, GUANINE NUCLEOTIDE EXCHANGE FACTOR DBS | | Authors: | Rossman, K.L, Worthylake, D.K, Snyder, J.T, Siderovski, D.P, Campbell, S.L, Sondek, J. | | Deposit date: | 2002-02-06 | | Release date: | 2002-03-20 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | A crystallographic view of interactions between Dbs and Cdc42: PH domain-assisted guanine nucleotide exchange.
EMBO J., 21, 2002
|
|
1KZ8
 
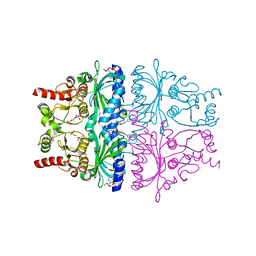 | | CRYSTAL STRUCTURE OF PORCINE FRUCTOSE-1,6-BISPHOSPHATASE COMPLEXED WITH A NOVEL ALLOSTERIC-SITE INHIBITOR | | Descriptor: | 6-O-phosphono-beta-D-fructofuranose, ADENOSINE MONOPHOSPHATE, FRUCTOSE-1,6-BISPHOSPHATASE, ... | | Authors: | Wright, S.W, Carlo, A.A, Carty, M.D, Danley, D.E, Hageman, D.L, Karam, G.A, Levy, C.B, Mansour, M.N, Mathiowetz, A.M, McClure, L.D, Nestor, N.B, McPherson, R.K, Pandit, J, Pustilnik, L.R, Schulte, G.K, Soeller, W.C, Treadway, J.L, Wang, I.-K, Bauer, P.H. | | Deposit date: | 2002-02-06 | | Release date: | 2002-10-16 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | ANILINOQUINAZOLINE INHIBITORS OF FRUCTOSE 1,6-BISPHOSPHATASE BIND AT A NOVEL ALLOSTERIC SITE: SYNTHESIS, IN VITRO CHARACTERIZATION, AND X-RAY CRYSTALLOGRAPHY
J.MED.CHEM., 45, 2002
|
|
1KZA
 
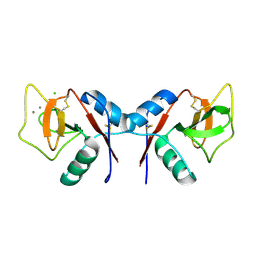 | | Complex of MBP-C and Man-a13-Man | | Descriptor: | CALCIUM ION, CHLORIDE ION, MANNOSE-BINDING PROTEIN C, ... | | Authors: | Ng, K.K, Kolatkar, A.R, Park-Snyder, S, Feinberg, H, Clark, D.A, Drickamer, K, Weis, W.I. | | Deposit date: | 2002-02-06 | | Release date: | 2002-07-05 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (1.74 Å) | | Cite: | Orientation of bound ligands in mannose-binding proteins. Implications for multivalent ligand recognition.
J.Biol.Chem., 277, 2002
|
|
1KZB
 
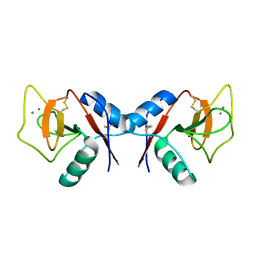 | | Complex of MBP-C and trimannosyl core | | Descriptor: | CALCIUM ION, MANNOSE-BINDING PROTEIN C, alpha-D-mannopyranose | | Authors: | Ng, K.K, Kolatkar, A.R, Park-Snyder, S, Feinberg, H, Clark, D.A, Drickamer, K, Weis, W.I. | | Deposit date: | 2002-02-06 | | Release date: | 2002-07-05 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Orientation of bound ligands in mannose-binding proteins. Implications for multivalent ligand recognition.
J.Biol.Chem., 277, 2002
|
|
1KZC
 
 | | Complex of MBP-C and high-affinity linear trimannose | | Descriptor: | CALCIUM ION, CHLORIDE ION, MANNOSE-BINDING PROTEIN C, ... | | Authors: | Ng, K.K, Kolatkar, A.R, Park-Snyder, S, Feinberg, H, Clark, D.A, Drickamer, K, Weis, W.I. | | Deposit date: | 2002-02-06 | | Release date: | 2002-07-05 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Orientation of bound ligands in mannose-binding proteins. Implications for multivalent ligand recognition.
J.Biol.Chem., 277, 2002
|
|
1KZD
 
 | | Complex of MBP-C and GlcNAc-terminated core | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, MANNOSE-BINDING PROTEIN C | | Authors: | Ng, K.K, Kolatkar, A.R, Park-Snyder, S, Feinberg, H, Clark, D.A, Drickamer, K, Weis, W.I. | | Deposit date: | 2002-02-06 | | Release date: | 2002-07-05 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Orientation of bound ligands in mannose-binding proteins. Implications for multivalent ligand recognition.
J.Biol.Chem., 277, 2002
|
|
