1G1J
 
 | |
1G1L
 
 | | THE STRUCTURAL BASIS OF THE CATALYTIC MECHANISM AND REGULATION OF GLUCOSE-1-PHOSPHATE THYMIDYLYLTRANSFERASE (RMLA). TDP-GLUCOSE COMPLEX. | | Descriptor: | 2'DEOXY-THYMIDINE-5'-DIPHOSPHO-ALPHA-D-GLUCOSE, CITRIC ACID, GLUCOSE-1-PHOSPHATE THYMIDYLYLTRANSFERASE, ... | | Authors: | Blankenfeldt, W, Asuncion, M, Lam, J.S, Naimsmith, J.H. | | Deposit date: | 2000-10-12 | | Release date: | 2000-12-27 | | Last modified: | 2018-01-31 | | Method: | X-RAY DIFFRACTION (1.77 Å) | | Cite: | The structural basis of the catalytic mechanism and regulation of glucose-1-phosphate thymidylyltransferase (RmlA).
EMBO J., 19, 2000
|
|
1G1U
 
 | | THE 2.5 ANGSTROM RESOLUTION CRYSTAL STRUCTURE OF THE RXRALPHA LIGAND BINDING DOMAIN IN TETRAMER IN THE ABSENCE OF LIGAND | | Descriptor: | RETINOIC ACID RECEPTOR RXR-ALPHA | | Authors: | Gampe Jr, R.T, Montana, V.G, Lambert, M.H, Wisely, G.B, Milburn, M.V, Xu, H.E. | | Deposit date: | 2000-10-13 | | Release date: | 2001-04-25 | | Last modified: | 2017-10-04 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structural basis for autorepression of retinoid X receptor by tetramer formation and the AF-2 helix.
Genes Dev., 14, 2000
|
|
1G1V
 
 | | T4 LYSOZYME MUTANT C54T/C97A/I58T | | Descriptor: | BETA-MERCAPTOETHANOL, CHLORIDE ION, LYSOZYME | | Authors: | Quillin, M.L, Matthews, B.W. | | Deposit date: | 2000-10-13 | | Release date: | 2001-05-09 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structural and thermodynamic analysis of the binding of solvent at internal sites in T4 lysozyme.
Protein Sci., 10, 2001
|
|
1G1W
 
 | | T4 LYSOZYME MUTANT C54T/C97A/Q105M | | Descriptor: | BETA-MERCAPTOETHANOL, CHLORIDE ION, LYSOZYME | | Authors: | Quillin, M.L, Matthews, B.W. | | Deposit date: | 2000-10-13 | | Release date: | 2001-05-09 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structural and thermodynamic analysis of the binding of solvent at internal sites in T4 lysozyme.
Protein Sci., 10, 2001
|
|
1G1Y
 
 | | CRYSTAL STRUCTURE OF ALPHA-AMYLASE II (TVAII) FROM THERMOACTINOMYCES VULGARIS R-47 AND BETA-CYCLODEXTRIN COMPLEX | | Descriptor: | ALPHA-AMYLASE II, Cycloheptakis-(1-4)-(alpha-D-glucopyranose) | | Authors: | Kondo, S, Ohtaki, A, Tonozuka, T, Sakano, Y, Kamitori, S. | | Deposit date: | 2000-10-16 | | Release date: | 2001-03-14 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Studies on the hydrolyzing mechanism for cyclodextrins of Thermoactinomyces vulgaris R-47 alpha-amylase 2 (TVAII). X-ray structure of the mutant E354A complexed with beta-cyclodextrin, and kinetic analyses on cyclodextrins.
J.Biochem.(Tokyo), 129, 2001
|
|
1G23
 
 | | THE STRUCTURAL BASIS OF THE CATALYTIC MECHANISM AND REGULATION OF GLUCOSE-1-PHOSPHATE THYMIDYLYLTRANSFERASE (RMLA). GLUCOSE-1-PHOSPHATE COMPLEX. | | Descriptor: | 1-O-phosphono-alpha-D-glucopyranose, GLUCOSE-1-PHOSPHATE THYMIDYLYLTRANSFERASE, SULFATE ION | | Authors: | Blankenfeldt, W, Asuncion, M, Lam, J.S, Naismith, J.H. | | Deposit date: | 2000-10-16 | | Release date: | 2000-12-27 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | The structural basis of the catalytic mechanism and regulation of glucose-1-phosphate thymidylyltransferase (RmlA).
EMBO J., 19, 2000
|
|
1G29
 
 | | MALK | | Descriptor: | 1,4-DIETHYLENE DIOXIDE, AMMONIUM ION, CHLORIDE ION, ... | | Authors: | Diederichs, K, Diez, J, Greller, G, Mueller, C, Breed, J, Schnell, C, Vonrhein, C, Boos, W, Welte, W. | | Deposit date: | 2000-10-18 | | Release date: | 2000-12-06 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal structure of MalK, the ATPase subunit of the trehalose/maltose ABC transporter of the archaeon Thermococcus litoralis.
EMBO J., 19, 2000
|
|
1G2B
 
 | | ALPHA-SPECTRIN SRC HOMOLOGY 3 DOMAIN, CIRCULAR PERMUTANT, CUT AT N47-D48 | | Descriptor: | SPECTRIN ALPHA CHAIN, SULFATE ION | | Authors: | Berisio, R, Viguera, A.R, Serrano, L, Wilmanns, M. | | Deposit date: | 2000-10-18 | | Release date: | 2000-11-01 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.12 Å) | | Cite: | Atomic resolution structure of a mutant of the spectrin SH3 domain.
Acta Crystallogr.,Sect.D, 57, 2001
|
|
1G2E
 
 | |
1G2I
 
 | | CRYSTAL STRUCTURE OF A NOVEL INTRACELLULAR PROTEASE FROM PYROCOCCUS HORIKOSHII AT 2 A RESOLUTION | | Descriptor: | PROTEASE I, SULFATE ION | | Authors: | Du, X, Choi, I.-G, Kim, R, Jancarik, J, Kim, S.-H, Berkeley Structural Genomics Center (BSGC) | | Deposit date: | 2000-10-19 | | Release date: | 2000-11-08 | | Last modified: | 2018-01-24 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal structure of an intracellular protease from Pyrococcus horikoshii at 2-A resolution.
Proc.Natl.Acad.Sci.USA, 97, 2000
|
|
1G2J
 
 | | RNA OCTAMER R(CCCP*GGGG) CONTAINING PHENYL RIBONUCLEOTIDE | | Descriptor: | 5'-R(*CP*CP*CP*(PYY)P*GP*GP*GP*G)-3', CALCIUM ION | | Authors: | Minasov, G, Matulic-Adamic, J, Wilds, C.J, Haeberli, P, Usman, N, Beigelman, L, Egli, M. | | Deposit date: | 2000-10-20 | | Release date: | 2000-12-06 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.97 Å) | | Cite: | Crystal structure of an RNA duplex containing phenyl-ribonucleotides, hydrophobic isosteres of the natural pyrimidines.
RNA, 6, 2000
|
|
1G2K
 
 | | HIV-1 PROTEASE WITH CYCLIC SULFAMIDE INHIBITOR, AHA047 | | Descriptor: | 3-(7-BENZYL-4,5-DIHYDROXY-1,1-DIOXO-3,6-BIS-PHENOXYMETHYL-1L6-[1,2,7]THIADIAZEPAN-2-YLMETHYL)-N-METHYL-BENZAMIDE, PROTEASE RETROPEPSIN | | Authors: | Lindberg, J, Unge, T. | | Deposit date: | 2000-10-20 | | Release date: | 2001-06-01 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Synthesis and comparative molecular field analysis (CoMFA) of symmetric and nonsymmetric cyclic sulfamide HIV-1 protease inhibitors.
J.Med.Chem., 44, 2001
|
|
1G2L
 
 | | FACTOR XA INHIBITOR COMPLEX | | Descriptor: | CALCIUM ION, COAGULATION FACTOR X, [(1-{2[(4-CARBAMIMIDOYL-PHENYLAMINO)-METHYL]-1-METHYL-1H-BENZOIMIDAZOL-5-YL}-CYCLOPROPYL)-PYRIDIN-2-YL-METHYLENEAMINOOXY]-ACETIC ACID ETHYL ESTER | | Authors: | Nar, H. | | Deposit date: | 2000-10-20 | | Release date: | 2001-10-20 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structural basis for inhibition promiscuity of dual specific thrombin and factor Xa blood coagulation inhibitors.
Structure, 9, 2001
|
|
1G2M
 
 | | FACTOR XA INHIBITOR COMPLEX | | Descriptor: | 4-{[1-METHYL-5-(2-METHYL-BENZOIMIDAZOL-1-YLMETHYL)-1H-BENZOIMIDAZOL-2-YLMETHYL]-AMINO}-BENZAMIDINE, CALCIUM ION, COAGULATION FACTOR X | | Authors: | Nar, H. | | Deposit date: | 2000-10-20 | | Release date: | 2001-10-20 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (3.02 Å) | | Cite: | Structural basis for inhibition promiscuity of dual specific thrombin and factor Xa blood coagulation inhibitors.
Structure, 9, 2001
|
|
1G2N
 
 | | CRYSTAL STRUCTURE OF THE LIGAND BINDING DOMAIN OF THE ULTRASPIRACLE PROTEIN USP, THE ORTHOLOG OF RXRS IN INSECTS | | Descriptor: | L-ALPHA-PHOSPHATIDYL-BETA-OLEOYL-GAMMA-PALMITOYL-PHOSPHATIDYLETHANOLAMINE, ULTRASPIRACLE PROTEIN | | Authors: | Billas, I.M.L, Moulinier, L, Rochel, N, Moras, D, Structural Proteomics in Europe (SPINE) | | Deposit date: | 2000-10-20 | | Release date: | 2001-04-21 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Crystal structure of the ligand-binding domain of the ultraspiracle protein USP, the ortholog of retinoid X receptors in insects.
J.Biol.Chem., 276, 2001
|
|
1G2R
 
 | | Structure of Cytosolic Protein of Unknown Function Coded by Gene from NUSA/INFB Region, a YlxR Homologue | | Descriptor: | HYPOTHETICAL CYTOSOLIC PROTEIN, SULFATE ION | | Authors: | Osipiuk, J, Gornicki, P, Maj, L, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2000-10-20 | | Release date: | 2001-08-29 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.35 Å) | | Cite: | Streptococcus pneumonia YlxR at 1.35 A shows a putative new fold.
Acta Crystallogr.,Sect.D, 57, 2001
|
|
1G2U
 
 | | THE STRUCTURE OF THE MUTANT, A172V, OF 3-ISOPROPYLMALATE DEHYDROGENASE FROM THERMUS THERMOPHILUS HB8 : ITS THERMOSTABILITY AND STRUCTURE. | | Descriptor: | 3-ISOPROPYLMALATE DEHYDROGENASE | | Authors: | Qu, C, Akanuma, S, Tanaka, N, Moriyama, H, Oshima, T. | | Deposit date: | 2000-10-21 | | Release date: | 2000-11-01 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Design, X-ray crystallography, molecular modelling and thermal stability studies of mutant enzymes at site 172 of 3-isopropylmalate dehydrogenase from Thermus thermophilus.
Acta Crystallogr.,Sect.D, 57, 2001
|
|
1G2V
 
 | | THE STRUCTURAL BASIS OF THE CATALYTIC MECHANISM AND REGULATION OF GLUCOSE-1-PHOSPHATE THYMIDYLYLTRANSFERASE (RMLA). TTP COMPLEX. | | Descriptor: | GLUCOSE-1-PHOSPHATE THYMIDYLYLTRANSFERASE, THYMIDINE-5'-TRIPHOSPHATE | | Authors: | Blankenfeldt, W, Asuncion, M, Lam, J.S, Naismith, J.H. | | Deposit date: | 2000-10-21 | | Release date: | 2000-12-27 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | The structural basis of the catalytic mechanism and regulation of glucose-1-phosphate thymidylyltransferase (RmlA).
EMBO J., 19, 2000
|
|
1G2W
 
 | | E177S MUTANT OF THE PYRIDOXAL-5'-PHOSPHATE ENZYME D-AMINO ACID AMINOTRANSFERASE | | Descriptor: | ACETATE ION, D-ALANINE AMINOTRANSFERASE, PYRIDOXAL-5'-PHOSPHATE | | Authors: | Lepore, B.W, Ringe, D. | | Deposit date: | 2000-10-21 | | Release date: | 2000-11-15 | | Last modified: | 2021-11-03 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Studies on an Active Site Residue, E177, That Affects Binding of the Coenzyme in D-Amino Acid Transaminase, and Mechanistic Studies on a Suicide Substrate
Biochemistry and Molecular Biology of Vitamin B6 and PQQ-dependent Proteins, 10th Annual International Symposium on Vitamin B6, 2000
|
|
1G2X
 
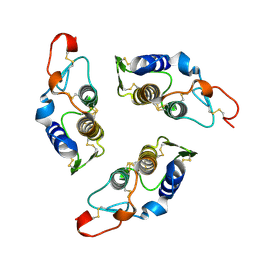 | | Sequence induced trimerization of krait PLA2: crystal structure of the trimeric form of krait PLA2 | | Descriptor: | PHOSPHOLIPASE A2 | | Authors: | Singh, G, Gourinath, S, Sharma, S, Bhanumathi, S, Paramsivam, M, Singh, T.P. | | Deposit date: | 2000-10-22 | | Release date: | 2003-06-17 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Sequence-induced trimerization of phospholipase A2: structure of a trimeric isoform of PLA2 from common krait (Bungarus caeruleus) at 2.5 A resolution.
Acta Crystallogr.,Sect.F, 61, 2005
|
|
1G2Y
 
 | | HNF-1ALPHA DIMERIZATION DOMAIN, WITH SELENOMETHIONINE SUBSTITUED AT LEU 12 | | Descriptor: | HEPATOCYTE NUCLEAR FACTOR 1-ALPHA | | Authors: | Rose, R.B, Endrizzi, J.A, Cronk, J.D, Holton, J, Alber, T. | | Deposit date: | 2000-10-23 | | Release date: | 2001-01-17 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1 Å) | | Cite: | High-resolution structure of the HNF-1alpha dimerization domain.
Biochemistry, 39, 2000
|
|
1G2Z
 
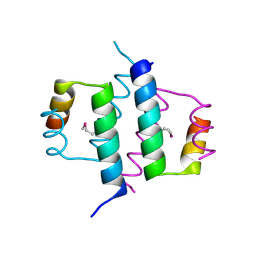 | | DIMERIZATION DOMAIN OF HNF-1ALPHA WITH A LEU 13 SELENOMETHIONINE SUBSTITUTION | | Descriptor: | HEPATOCYTE NUCLEAR FACTOR 1-ALPHA | | Authors: | Rose, R.B, Endrizzi, J.A, Cronk, J.D, Holton, J, Alber, T. | | Deposit date: | 2000-10-23 | | Release date: | 2001-01-17 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.15 Å) | | Cite: | High-resolution structure of the HNF-1alpha dimerization domain.
Biochemistry, 39, 2000
|
|
1G30
 
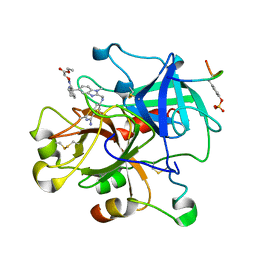 | | THROMBIN INHIBITOR COMPLEX | | Descriptor: | HIRUDIN IIB, PROTHROMBIN, [(1-{2[(4-CARBAMIMIDOYL-PHENYLAMINO)-METHYL]-1-METHYL-1H-BENZOIMIDAZOL-5-YL}-CYCLOPROPYL)-PYRIDIN-2-YL-METHYLENEAMINOOXY]-ACETIC ACID ETHYL ESTER | | Authors: | Nar, H. | | Deposit date: | 2000-10-23 | | Release date: | 2001-10-23 | | Last modified: | 2018-10-03 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural basis for inhibition promiscuity of dual specific thrombin and factor Xa blood coagulation inhibitors.
Structure, 9, 2001
|
|
1G31
 
 | | GP31 CO-CHAPERONIN FROM BACTERIOPHAGE T4 | | Descriptor: | GP31, PHOSPHATE ION, POTASSIUM ION | | Authors: | Hunt, J.F, Van Der Vies, S.M, Henry, L, Deisenhofer, J. | | Deposit date: | 1998-03-27 | | Release date: | 1998-08-26 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structural adaptations in the specialized bacteriophage T4 co-chaperonin Gp31 expand the size of the Anfinsen cage.
Cell(Cambridge,Mass.), 90, 1997
|
|
