1PYU
 
 | | Processed Aspartate Decarboxylase Mutant with Ser25 mutated to Cys | | Descriptor: | Aspartate 1-decarboxylase alfa chain, Aspartate 1-decarboxylase beta chain, SULFATE ION | | Authors: | Schmitzberger, F, Kilkenny, M.L, Lobley, C.M.C, Webb, M.E, Vinkovic, M, Matak-Vinkovic, D, Witty, M, Chirgadze, D.Y, Smith, A.G, Abell, C, Blundell, T.L. | | Deposit date: | 2003-07-09 | | Release date: | 2003-11-18 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structural Constraints on protein self-processing in L-aspartate-alpha-decarboxylase
Embo J., 22, 2003
|
|
1PYW
 
 | | Human class II MHC protein HLA-DR1 bound to a designed peptide related to influenza virus hemagglutinin, FVKQNA(MAA)AL, in complex with staphylococcal enterotoxin C3 variant 3B2 (SEC3-3B2) | | Descriptor: | 9-residue influenza virus hemagglutinin related peptide FVKQNA(MAA)AL, Enterotoxin type C-3, HLA class II histocompatibility antigen, ... | | Authors: | Zavala-Ruiz, Z, Sundberg, E.J, Stone, J.D, DeOliveira, D.B, Chan, I.C, Svendsen, J, Mariuzza, R.A, Stern, L.J. | | Deposit date: | 2003-07-09 | | Release date: | 2003-12-09 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Exploration of the P6/P7 region of the peptide-binding site of the human class II Major Histocompatability Complex Protein HLA-DR1
J.Biol.Chem., 278, 2003
|
|
1PYX
 
 | | GSK-3 Beta complexed with AMP-PNP | | Descriptor: | Glycogen synthase kinase-3 beta, MAGNESIUM ION, PHOSPHOAMINOPHOSPHONIC ACID-ADENYLATE ESTER | | Authors: | Bertrand, J.A, Thieffine, S, Vulpetti, A, Cristiani, C, Valsasina, B, Knapp, S, Kalisz, H.M, Flocco, M. | | Deposit date: | 2003-07-09 | | Release date: | 2003-10-21 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structural characterization of the GSK-3beta active site using selective and non-selective ATP-mimetic inhibitors
J.Mol.Biol., 333, 2003
|
|
1PYY
 
 | | Double mutant PBP2x T338A/M339F from Streptococcus pneumoniae strain R6 at 2.4 A resolution | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, 6-O-octanoyl-beta-D-fructofuranose-(2-1)-alpha-D-glucopyranose, Penicillin-binding protein 2X, ... | | Authors: | Chesnel, L, Pernot, L, Lemaire, D, Champelovier, D, Croize, J, Dideberg, O, Vernet, T, Zapun, A. | | Deposit date: | 2003-07-09 | | Release date: | 2003-09-30 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2.42 Å) | | Cite: | The Structural Modifications Induced by the M339F Substitution in PBP2x from Streptococcus pneumoniae Further Decreases the Susceptibility to beta-Lactams of Resistant Strains
J.Biol.Chem., 278, 2003
|
|
1PYZ
 
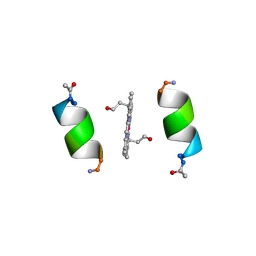 | | CRYSTALLOGRAPHIC STRUCTURE OF MIMOCHROME IV | | Descriptor: | CHLORIDE ION, CO(III)-(DEUTEROPORPHYRIN IX), MIMOCHROME IV, ... | | Authors: | Di Costanzo, L, Geremia, S, Randaccio, L, Nastri, F, Maglio, O, Lombardi, A, Pavone, V. | | Deposit date: | 2003-07-09 | | Release date: | 2004-12-14 | | Last modified: | 2018-06-27 | | Method: | X-RAY DIFFRACTION (1.25 Å) | | Cite: | Miniaturized heme proteins: crystal structure of Co(III)-mimochrome IV.
J.Biol.Inorg.Chem., 9, 2004
|
|
1PZ0
 
 | |
1PZ1
 
 | |
1PZ2
 
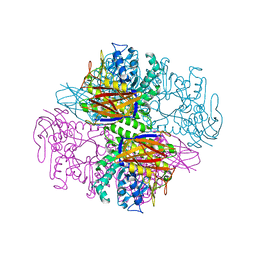 | | Crystal structure of a transient covalent reaction intermediate of a family 51 alpha-L-arabinofuranosidase | | Descriptor: | Alpha-L-arabinofuranosidase, alpha-L-arabinofuranose | | Authors: | Hoevel, K, Shallom, D, Niefind, K, Belakhov, V, Shoham, G, Baasov, T, Shoham, Y, Schomburg, D. | | Deposit date: | 2003-07-09 | | Release date: | 2003-10-07 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal structure and snapshots along the reaction pathway of a family 51 alpha-L-arabinofuranosidase
Embo J., 22, 2003
|
|
1PZ3
 
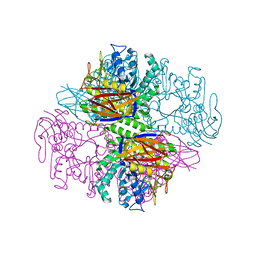 | | Crystal structure of a family 51 (GH51) alpha-L-arabinofuranosidase from Geobacillus stearothermophilus T6 | | Descriptor: | Alpha-L-arabinofuranosidase, GLYCEROL | | Authors: | Hoevel, K, Shallom, D, Niefind, K, Belakhov, V, Shoham, G, Baasov, T, Shoham, Y, Schomburg, D. | | Deposit date: | 2003-07-09 | | Release date: | 2003-10-07 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Crystal structure and snapshots along the reaction pathway of a family 51 alpha-L-arabinofuranosidase
Embo J., 22, 2003
|
|
1PZ4
 
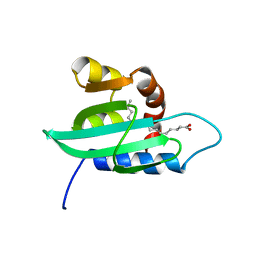 | | The structural determination of an insect (mosquito) Sterol Carrier Protein-2 with a ligand bound C16 Fatty Acid at 1.35 A resolution | | Descriptor: | PALMITIC ACID, sterol carrier protein 2 | | Authors: | Dyer, D.H, Lovell, S, Thoden, J.B, Holden, H.M, Rayment, I, Lan, Q. | | Deposit date: | 2003-07-09 | | Release date: | 2003-09-30 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.35 Å) | | Cite: | The Structural Determination of an Insect Sterol Carrier Protein-2 with a Ligand-bound C16 Fatty Acid at 1.35A Resolution
J.Biol.Chem., 278, 2003
|
|
1PZA
 
 | |
1PZB
 
 | |
1PZC
 
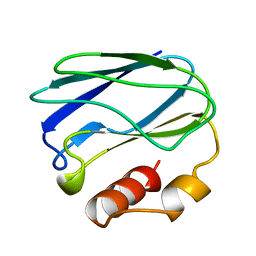 | | APO-PSEUDOAZURIN (METAL FREE PROTEIN) | | Descriptor: | PSEUDOAZURIN | | Authors: | Petratos, K. | | Deposit date: | 1995-02-22 | | Release date: | 1995-09-15 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | The crystal structure of apo-pseudoazurin from Alcaligenes faecalis S-6.
Febs Lett., 368, 1995
|
|
1PZD
 
 | | Structural Identification of a conserved appendage domain in the carboxyl-terminus of the COPI gamma-subunit. | | Descriptor: | Coatomer gamma subunit, SULFATE ION | | Authors: | Hoffman, G.R, Rahl, P.B, Collins, R.N, Cerione, R.A. | | Deposit date: | 2003-07-10 | | Release date: | 2003-10-14 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.31 Å) | | Cite: | Conserved Structural Motifs in Intracellular Trafficking Pathways. Structure of the gammaCOP Appendage Domain.
Mol.Cell, 12, 2003
|
|
1PZE
 
 | | T.gondii LDH1 apo form | | Descriptor: | lactate dehydrogenase | | Authors: | Kavanagh, K.L, Elling, R.A, Wilson, D.K. | | Deposit date: | 2003-07-10 | | Release date: | 2004-02-24 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Structure of Toxoplasma gondii LDH1: Active-Site Differences from Human Lactate Dehydrogenases and the Structural Basis for Efficient APAD+ Use.
Biochemistry, 43, 2004
|
|
1PZF
 
 | | T.gondii LDH1 ternary complex with APAD+ and oxalate | | Descriptor: | 3-ACETYLPYRIDINE ADENINE DINUCLEOTIDE, OXALATE ION, lactate dehydrogenase | | Authors: | Kavanagh, K.L, Elling, R.A, Wilson, D.K. | | Deposit date: | 2003-07-10 | | Release date: | 2004-02-24 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structure of Toxoplasma gondii LDH1: Active-Site Differences from Human Lactate Dehydrogenases and the Structural Basis for Efficient APAD+ Use.
Biochemistry, 43, 2004
|
|
1PZG
 
 | |
1PZH
 
 | | T.gondii LDH1 ternary complex with NAD and oxalate | | Descriptor: | NICOTINAMIDE-ADENINE-DINUCLEOTIDE, OXALATE ION, lactate dehydrogenase | | Authors: | Kavanagh, K.L, Elling, R.A, Wilson, D.K. | | Deposit date: | 2003-07-10 | | Release date: | 2004-02-24 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structure of Toxoplasma gondii LDH1: Active-Site Differences from Human Lactate Dehydrogenases and the Structural Basis for Efficient APAD+ Use.
Biochemistry, 43, 2004
|
|
1PZI
 
 | | Heat-Labile Enterotoxin B-Pentamer Complexed With Nitrophenyl Galactoside 2a | | Descriptor: | Heat-labile Enterotoxin B subunit, N-(2-MORPHOLIN-4-YL-1-MORPHOLIN-4-YLMETHYL-ETHYL)-3-NITRO-5-(3,4,5-TRIHYDROXY-6-HYDROXYMETHYL-TETRAHYDRO-PYRAN-2-YLOXY)-BENZAMIDE | | Authors: | Mitchell, D.D, Pickens, J.C, Korotkov, K, Fan, E, Hol, W.G.J. | | Deposit date: | 2003-07-11 | | Release date: | 2004-03-09 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (1.99 Å) | | Cite: | 3,5-Substituted phenyl galactosides as leads in designing effective cholera toxin antagonists; synthesis and crystallographic studies
Bioorg.Med.Chem., 12, 2004
|
|
1PZJ
 
 | | Cholera Toxin B-Pentamer Complexed With Nitrophenyl Galactoside 5 | | Descriptor: | Cholera Toxin B Subunit, N-{3-[4-(3-AMINO-PROPYL)-PIPERAZIN-1-YL]-PROPYL}-3-NITRO-5-(GALACTOPYRANOSYL)-ALPHA-BENZAMIDE, N-{3-[4-(3-AMINO-PROPYL)-PIPERAZIN-1-YL]-PROPYL}-3-NITRO-5-(GALACTOPYRANOSYL)-BETA-BENZAMIDE | | Authors: | Mitchell, D.D, Pickens, J.C, Korotkov, K, Fan, E, Hol, W.G.J. | | Deposit date: | 2003-07-11 | | Release date: | 2004-03-09 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (1.46 Å) | | Cite: | 3,5-Substituted phenyl galactosides as leads in designing effective cholera toxin antagonists; synthesis and crystallographic studies
Bioorg.Med.Chem., 12, 2004
|
|
1PZK
 
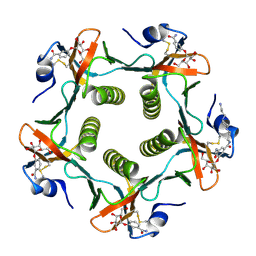 | | Cholera Toxin B-Pentamer Complexed With N-Acyl Phenyl Galactoside 9h | | Descriptor: | Cholera Toxin B Subunit, N-{3-[4-(3-AMINO-PROPYL)-PIPERAZIN-1-YL]-PROPYL}-3-(2-THIOPHEN-2-YL-ACETYLAMINO)-5-(3,4,5-TRIHYDROXY-6-HYDROXYMETHYL-TETRAHYDRO-PYRAN-2-YLOXY)-BENZAMIDE | | Authors: | Mitchell, D.D, Pickens, J.C, Korotkov, K, Fan, E, Hol, W.G.J. | | Deposit date: | 2003-07-11 | | Release date: | 2004-03-09 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (1.35 Å) | | Cite: | 3,5-Substituted phenyl galactosides as leads in designing effective cholera toxin antagonists; synthesis and crystallographic studies
Bioorg.Med.Chem., 12, 2004
|
|
1PZL
 
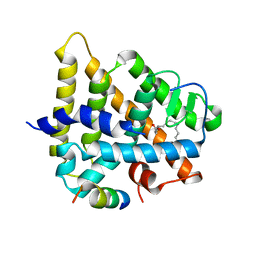 | | Crystal structure of HNF4a LBD in complex with the ligand and the coactivator SRC-1 peptide | | Descriptor: | Hepatocyte nuclear factor 4-alpha, MYRISTIC ACID, steroid receptor coactivator-1 | | Authors: | Duda, K, Chi, Y.-I, Dhe-paganon, S, Shoelson, S. | | Deposit date: | 2003-07-11 | | Release date: | 2004-06-01 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structural Basis for HNF-4alpha Activation by Ligand and Coactivator Binding
J.Biol.Chem., 279, 2004
|
|
1PZM
 
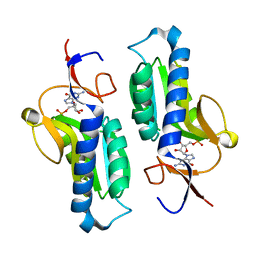 | | Crystal structure of HGPRT-ase from Leishmania tarentolae in complex with GMP | | Descriptor: | GUANOSINE-5'-MONOPHOSPHATE, hypoxanthine-guanine phosphoribosyltransferase | | Authors: | Monzani, P.S, Trapani, S, Oliva, G, Thiemann, O.H. | | Deposit date: | 2003-07-11 | | Release date: | 2004-07-27 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Crystal structure of Leishmania tarentolae hypoxanthine-guanine phosphoribosyltransferase.
Bmc Struct.Biol., 7, 2007
|
|
1PZN
 
 | | Rad51 (RadA) | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, DNA repair and recombination protein rad51, GLYCEROL, ... | | Authors: | Shin, D.S, Tainer, J.A. | | Deposit date: | 2003-07-12 | | Release date: | 2003-09-09 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.85 Å) | | Cite: | Full-length archaeal Rad51 structure and mutants: Mechanisms for RAD51 assembly and control by BRCA2
Embo J., 22, 2003
|
|
1PZO
 
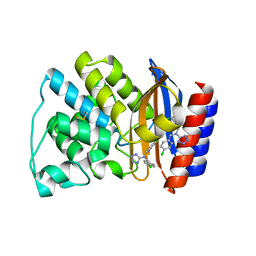 | | TEM-1 Beta-Lactamase in Complex with a Novel, Core-Disrupting, Allosteric Inhibitor | | Descriptor: | Beta-lactamase TEM, N,N-BIS(4-CHLOROBENZYL)-1H-1,2,3,4-TETRAAZOL-5-AMINE | | Authors: | Horn, J.R, Shoichet, B.K. | | Deposit date: | 2003-07-14 | | Release date: | 2004-03-09 | | Last modified: | 2021-10-27 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Allosteric inhibition through core disruption.
J.Mol.Biol., 336, 2004
|
|
