1MWQ
 
 | | Structure of HI0828, a Hypothetical Protein from Haemophilus influenzae with a Putative Active-Site Phosphohistidine | | Descriptor: | CACODYLATE ION, CHLORIDE ION, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Willis, M.A, Krajewski, W, Chalamasetty, V.R, Reddy, P, Howard, A, Herzberg, O, Structure 2 Function Project (S2F) | | Deposit date: | 2002-09-30 | | Release date: | 2003-11-25 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (0.99 Å) | | Cite: | Structure of YciI from Haemophilus influenzae (HI0828) reveals a ferredoxin-like alpha/beta-fold with a histidine/aspartate centered catalytic site
Proteins, 59, 2005
|
|
1MWR
 
 | |
1MWS
 
 | |
1MWT
 
 | |
1MWU
 
 | | Structure of methicillin acyl-Penicillin binding protein 2a from methicillin resistant Staphylococcus aureus strain 27r at 2.60 A resolution. | | Descriptor: | (2R,4S)-2-[(1R)-1-{[(2,6-dimethoxyphenyl)carbonyl]amino}-2-oxoethyl]-5,5-dimethyl-1,3-thiazolidine-4-carboxylic acid, CADMIUM ION, CHLORIDE ION, ... | | Authors: | Lim, D.C, Strynadka, N.C.J. | | Deposit date: | 2002-10-01 | | Release date: | 2002-11-06 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Structural basis for the beta lactam resistance of PBP2a from methicillin-resistant Staphylococcus aureus.
Nat.Struct.Biol., 9, 2002
|
|
1MWW
 
 | | THE STRUCTURE OF THE HYPOTHETICAL PROTEIN HI1388.1 FROM HAEMOPHILUS INFLUENZAE REVEALS A TAUTOMERASE/MIF FOLD | | Descriptor: | CHLORIDE ION, GLUTAMIC ACID, HYPOTHETICAL PROTEIN HI1388.1 | | Authors: | Lehmann, C, Pullalarevu, S, Krajewski, W, Galkin, A, Howard, A, Herzberg, O, Structure 2 Function Project (S2F) | | Deposit date: | 2002-10-01 | | Release date: | 2003-11-18 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.08 Å) | | Cite: | Structure of the Hypothetical Protein HI1388.1 from Haemophilus influenzae
To be Published
|
|
1MX0
 
 | | Structure of topoisomerase subunit | | Descriptor: | MAGNESIUM ION, PHOSPHOAMINOPHOSPHONIC ACID-ADENYLATE ESTER, SODIUM ION, ... | | Authors: | Corbett, K.D, Berger, J.M. | | Deposit date: | 2002-10-01 | | Release date: | 2003-01-07 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structure of the topoisomerase VI-B subunit: implications for type II topoisomerase mechanism and evolution
Embo J., 22, 2003
|
|
1MX3
 
 | | Crystal structure of CtBP dehydrogenase core holo form | | Descriptor: | ACETIC ACID, C-terminal binding protein 1, NICOTINAMIDE-ADENINE-DINUCLEOTIDE | | Authors: | Kumar, V, Carlson, J.E, Ohgi, K.E, Edwards, T.E, Rose, D.W, Escalante, C.R, Aggarwal, A.K. | | Deposit date: | 2002-10-01 | | Release date: | 2002-12-18 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Transcription Corepressor CtBP Is an NAD+-Regulated Dehydrogenase
Mol.Cell, 10, 2002
|
|
1MXD
 
 | | Structure of a (Ca,Zn)-dependent alpha-amylase from the hyperthermophilic archaeon Pyrococcus woesei | | Descriptor: | 4,6-dideoxy-4-{[(1S,4R,5S,6S)-4,5,6-trihydroxy-3-(hydroxymethyl)cyclohex-2-en-1-yl]amino}-alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose, ACETATE ION, CALCIUM ION, ... | | Authors: | Linden, A, Mayans, O, Meyer-Klaucke, W, Antranikian, G, Wilmanns, M. | | Deposit date: | 2002-10-02 | | Release date: | 2003-06-10 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Differential Regulation of a Hyperthermophilic alpha-Amylase with a Novel (Ca,Zn) Two-metal Center by Zinc
J.Biol.Chem., 278, 2003
|
|
1MXE
 
 | | Structure of the Complex of Calmodulin with the Target Sequence of CaMKI | | Descriptor: | CALCIUM ION, Calmodulin, Target Sequence of rat Calmodulin-Dependent Protein Kinase I | | Authors: | Clapperton, J.A, Martin, S.R, Smerdon, S.J, Gamblin, S.J, Bayley, P.M. | | Deposit date: | 2002-10-02 | | Release date: | 2002-12-04 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structure of the Complex
of Calmodulin with the Target
Sequence of Calmodulin-Dependent
Protein Kinase I: Studies of the
Kinase Activation Mechanism
Biochemistry, 41, 2002
|
|
1MXF
 
 | | Crystal Structure of Inhibitor Complex of Putative Pteridine Reductase 2 (PTR2) from Trypanosoma cruzi | | Descriptor: | METHOTREXATE, NADPH DIHYDRO-NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, PTERIDINE REDUCTASE 2 | | Authors: | Schormann, N, Pal, B, Senkovich, O, Carson, M, Howard, A, Smith, C, Delucas, L, Chattopadhyay, D. | | Deposit date: | 2002-10-02 | | Release date: | 2003-10-14 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystal structure of Trypanosoma cruzi pteridine reductase 2 in complex with a substrate and an inhibitor.
J.Struct.Biol., 152, 2005
|
|
1MXG
 
 | | Crystal Structure of a (Ca,Zn)-dependent alpha-amylase from the hyperthermophilic archaeon Pyrococcus woesei in complex with acarbose | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, 2-{2-[2-2-(METHOXY-ETHOXY)-ETHOXY]-ETHOXY}-ETHANOL, 4,6-dideoxy-4-{[(1S,4R,5S,6S)-4,5,6-trihydroxy-3-(hydroxymethyl)cyclohex-2-en-1-yl]amino}-alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose, ... | | Authors: | Linden, A, Mayans, O, Meyer-Klaucke, W, Antranikian, G, Wilmanns, M. | | Deposit date: | 2002-10-02 | | Release date: | 2003-06-10 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Differential Regulation of a Hyperthermophilic alpha-Amylase with a Novel (Ca,Zn) Two-metal Center by Zinc
J.Biol.Chem., 278, 2003
|
|
1MXH
 
 | | Crystal Structure of Substrate Complex of Putative Pteridine Reductase 2 (PTR2) from Trypanosoma cruzi | | Descriptor: | DIHYDROFOLIC ACID, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, PTERIDINE REDUCTASE 2 | | Authors: | Schormann, N, Pal, B, Senkovich, O, Carson, M, Howard, A, Smith, C, Delucas, L, Chattopadhyay, D. | | Deposit date: | 2002-10-02 | | Release date: | 2003-10-14 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystal structure of Trypanosoma cruzi pteridine reductase 2 in complex with a substrate and an inhibitor.
J.Struct.Biol., 152, 2005
|
|
1MXI
 
 | | Structure of YibK from Haemophilus influenzae (HI0766): a Methyltransferase with a Cofactor Bound at a Site Formed by a Knot | | Descriptor: | Hypothetical tRNA/rRNA methyltransferase HI0766, IODIDE ION, S-ADENOSYL-L-HOMOCYSTEINE | | Authors: | Lim, K, Zhang, H, Tempczyk, A, Bonander, N, Toedt, J, Howard, A, Eisenstein, E, Herzberg, O, Structure 2 Function Project (S2F) | | Deposit date: | 2002-10-02 | | Release date: | 2003-02-25 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structure of the YibK methyltransferase from Haemophilus influenzae (HI0766): a Cofactor Bound at a Site Formed by a Knot
Proteins, 51, 2003
|
|
1MXO
 
 | | AmpC beta-lactamase in complex with an m.carboxyphenylglycylboronic acid bearing the cephalothin R1 side chain | | Descriptor: | (1R)-1-(2-THIENYLACETYLAMINO)-1-(3-CARBOXYPHENYL)METHYLBORONIC ACID, Beta-lactamase, PHOSPHATE ION | | Authors: | Morandi, F, Caselli, E, Morandi, S, Focia, P.J, Blazquez, J, Shoichet, B.K, Prati, F. | | Deposit date: | 2002-10-02 | | Release date: | 2003-03-04 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.83 Å) | | Cite: | Nanomolar inhibitors of AmpC beta-lactamase.
J.Am.Chem.Soc., 125, 2003
|
|
1MXR
 
 | | High resolution structure of Ribonucleotide reductase R2 from E. coli in its oxidised (Met) form | | Descriptor: | FE (III) ION, GLYCEROL, MERCURY (II) ION, ... | | Authors: | Andersson, M.A, Hogbom, M, Nordlund, P. | | Deposit date: | 2002-10-03 | | Release date: | 2003-03-25 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.42 Å) | | Cite: | Displacement of the tyrosyl radical cofactor in ribonucleotide reductase obtained by single-crystal high-field EPR and 1.4-A x-ray data.
Proc.Natl.Acad.Sci.Usa, 100, 2003
|
|
1MXS
 
 | | Crystal structure of 2-keto-3-deoxy-6-phosphogluconate (KDPG) aldolase from Pseudomonas putida. | | Descriptor: | KDPG Aldolase, SULFATE ION | | Authors: | Watanabe, L, Bell, B.J, Lebioda, L, Rios-Steiner, J.L, Tulinsky, A, Arni, R.K. | | Deposit date: | 2002-10-03 | | Release date: | 2003-09-16 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structure of 2-keto-3-deoxy-6-phosphogluconate (KDPG) aldolase from Pseudomonas putida.
Acta Crystallogr.,Sect.D, 59, 2003
|
|
1MY6
 
 | | The 1.6 A Structure of Fe-Superoxide Dismutase from the thermophilic cyanobacterium Thermosynechococcus elongatus : Correlation of EPR and Structural Characteristics | | Descriptor: | FE (III) ION, Iron (III) Superoxide Dismutase | | Authors: | Yoshida, S.M, Cascio, D, Sawaya, M.R, Yeates, T.O, Kerfeld, C.A. | | Deposit date: | 2002-10-03 | | Release date: | 2003-07-29 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | The 1.6 A resolution structure of Fe-superoxide dismutase from the thermophilic cyanobacterium Thermosynechococcus elongatus.
J.BIOL.INORG.CHEM., 8, 2003
|
|
1MY8
 
 | | AmpC beta-lactamase in complex with an M.carboxyphenylglycylboronic acid bearing the cephalothin R1 side chain | | Descriptor: | (1R)-1-(2-THIENYLACETYLAMINO)-1-PHENYLMETHYLBORONIC ACID, PHOSPHATE ION, beta-lactamase | | Authors: | Morandi, F, Caselli, E, Morandi, S, Focia, P.J, Blazquez, J, Shoichet, B.K, Prati, F. | | Deposit date: | 2002-10-03 | | Release date: | 2003-03-04 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.72 Å) | | Cite: | Nanomolar inhibitors of AmpC beta-lactamase.
J.Am.Chem.Soc., 125, 2003
|
|
1MYG
 
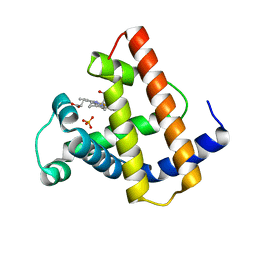 | | HIGH RESOLUTION X-RAY STRUCTURES OF PIG METMYOGLOBIN AND TWO CD3 MUTANTS MB(LYS45-> ARG) AND MB(LYS45-> SER) | | Descriptor: | MYOGLOBIN, PROTOPORPHYRIN IX CONTAINING FE, SULFATE ION | | Authors: | Smerdon, S.J, Oldfield, T.J, Wilkinson, A.J, Dauter, Z, Petratos, K, Wilson, K.S. | | Deposit date: | 1992-02-27 | | Release date: | 1994-01-31 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | High-resolution X-ray structures of pig metmyoglobin and two CD3 mutants: Mb(Lys45----Arg) and Mb(Lys45----Ser).
Biochemistry, 31, 1992
|
|
1MYH
 
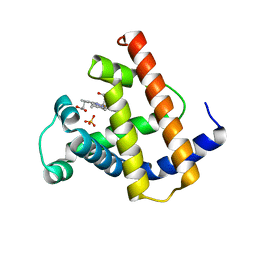 | | HIGH RESOLUTION X-RAY STRUCTURES OF PIG METMYOGLOBIN AND TWO CD3 MUTANTS MB(LYS45-> ARG) AND MB(LYS45-> SER) | | Descriptor: | MYOGLOBIN, PROTOPORPHYRIN IX CONTAINING FE, SULFATE ION | | Authors: | Smerdon, S.J, Oldfield, T.J, Wilkinson, A.J, Dauter, Z, Petratos, K, Wilson, K.S. | | Deposit date: | 1992-02-27 | | Release date: | 1994-01-31 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | High-resolution X-ray structures of pig metmyoglobin and two CD3 mutants: Mb(Lys45----Arg) and Mb(Lys45----Ser).
Biochemistry, 31, 1992
|
|
1MYI
 
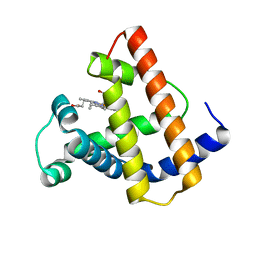 | | HIGH RESOLUTION X-RAY STRUCTURES OF PIG METMYOGLOBIN AND TWO CD3 MUTANTS MB(LYS45-> ARG) AND MB(LYS45-> SER) | | Descriptor: | MYOGLOBIN, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Smerdon, S.J, Oldfield, T.J, Wilkinson, A.J, Dauter, Z, Petratos, K, Wilson, K.S. | | Deposit date: | 1992-02-27 | | Release date: | 1994-01-31 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | High-resolution X-ray structures of pig metmyoglobin and two CD3 mutants: Mb(Lys45----Arg) and Mb(Lys45----Ser).
Biochemistry, 31, 1992
|
|
1MYJ
 
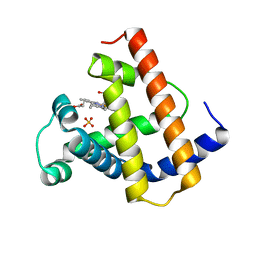 | | DISTAL POLARITY IN LIGAND BINDING TO MYOGLOBIN: STRUCTURAL AND FUNCTIONAL CHARACTERIZATION OF A THREONINE68(E11) MUTANT | | Descriptor: | MYOGLOBIN, PROTOPORPHYRIN IX CONTAINING FE, SULFATE ION | | Authors: | Smerdon, S.J, Oldfield, T.J, Wilkinson, A.J, Dauter, Z, Petratos, K, Wilson, K.S. | | Deposit date: | 1992-02-27 | | Release date: | 1994-01-31 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Distal pocket polarity in ligand binding to myoglobin: structural and functional characterization of a threonine68(E11) mutant.
Biochemistry, 30, 1991
|
|
1MYM
 
 | |
1MYR
 
 | | MYROSINASE FROM SINAPIS ALBA | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, GLYCEROL, ... | | Authors: | Burmeister, W.P, Iori, R, Palmieri, S, Henrissat, B. | | Deposit date: | 1997-03-23 | | Release date: | 1997-06-16 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.64 Å) | | Cite: | The crystal structures of Sinapis alba myrosinase and a covalent glycosyl-enzyme intermediate provide insights into the substrate recognition and active-site machinery of an S-glycosidase.
Structure, 5, 1997
|
|
