1LOG
 
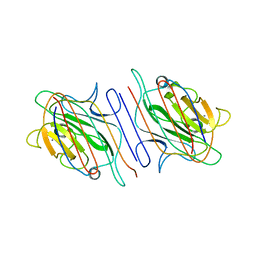 | |
1LOH
 
 | | Streptococcus pneumoniae Hyaluronate Lyase in Complex with Hexasaccharide Hyaluronan Substrate | | Descriptor: | Hyaluronate Lyase, beta-D-glucopyranuronic acid-(1-3)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-beta-D-glucopyranuronic acid-(1-3)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-beta-D-glucopyranuronic acid-(1-3)-2-acetamido-2-deoxy-beta-D-glucopyranose | | Authors: | Jedrzejas, M.J, Mello, L.V, De Groot, B.L, Li, S. | | Deposit date: | 2002-05-06 | | Release date: | 2002-08-07 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Mechanism of hyaluronan degradation by Streptococcus pneumoniae hyaluronate lyase. Structures of complexes with the substrate.
J.Biol.Chem., 277, 2002
|
|
1LOJ
 
 | | Crystal structure of a Methanobacterial Sm-like archaeal protein (SmAP1) bound to uridine-5'-monophosphate (UMP) | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, URIDINE, URIDINE-5'-MONOPHOSPHATE, ... | | Authors: | Mura, C, Kozhukhovsky, A, Eisenberg, D. | | Deposit date: | 2002-05-06 | | Release date: | 2003-03-25 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | The oligomerization and ligand-binding properties of Sm-like archaeal proteins (SmAPs)
Protein Sci., 12, 2003
|
|
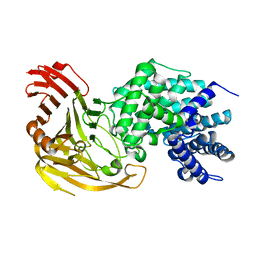
1LOL
 
 | | Crystal structure of orotidine monophosphate decarboxylase complex with XMP | | Descriptor: | 1,3-BUTANEDIOL, XANTHOSINE-5'-MONOPHOSPHATE, orotidine 5'-monophosphate decarboxylase | | Authors: | Wu, N, Pai, E.F. | | Deposit date: | 2002-05-06 | | Release date: | 2002-08-07 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal structures of inhibitor complexes reveal an alternate binding mode in orotidine-5'-monophosphate decarboxylase.
J.Biol.Chem., 277, 2002
|
|
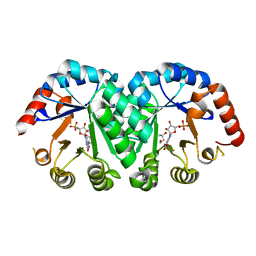
1LOM
 
 | | CYANOVIRIN-N DOUBLE MUTANT P51S S52P | | Descriptor: | CALCIUM ION, Cyanovirin-N, SULFATE ION | | Authors: | Botos, I, Mori, T, Cartner, L.K, Boyd, M.R, Wlodawer, A. | | Deposit date: | 2002-05-06 | | Release date: | 2002-06-26 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (1.72 Å) | | Cite: | Domain-swapped structure of a mutant of cyanovirin-N.
Biochem.Biophys.Res.Commun., 294, 2002
|
|
1LON
 
 | | Crystal Structure of the Recombinant Mouse-Muscle Adenylosuccinate Synthetase Complexed with 6-phosphoryl-IMP, GDP and Hadacidin | | Descriptor: | 6-O-PHOSPHORYL INOSINE MONOPHOSPHATE, GUANOSINE-5'-DIPHOSPHATE, HADACIDIN, ... | | Authors: | Iancu, C.V, Borza, T, Fromm, H.J, Honzatko, R.B. | | Deposit date: | 2002-05-06 | | Release date: | 2002-08-28 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | IMP, GTP, and 6-phosphoryl-IMP complexes of recombinant mouse muscle adenylosuccinate synthetase.
J.Biol.Chem., 277, 2002
|
|
1LOO
 
 | | Crystal Structure of the Mouse-Muscle Adenylosuccinate Synthetase Ligated with GTP | | Descriptor: | GUANOSINE-5'-TRIPHOSPHATE, adenylosuccinate synthetase | | Authors: | Iancu, C.V, Borza, T, Fromm, H.J, Honzatko, R.B. | | Deposit date: | 2002-05-06 | | Release date: | 2002-08-28 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | IMP, GTP, and 6-phosphoryl-IMP complexes of recombinant mouse muscle adenylosuccinate synthetase.
J.Biol.Chem., 277, 2002
|
|
1LOP
 
 | | CYCLOPHILIN A COMPLEXED WITH SUCCINYL-ALA-PRO-ALA-P-NITROANILIDE | | Descriptor: | CYCLOPHILIN A, SUCCINYL-ALA-PRO-ALA-P-NITROANILIDE | | Authors: | Konno, M. | | Deposit date: | 1996-06-17 | | Release date: | 1996-12-23 | | Last modified: | 2017-11-29 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | The substrate-binding site in Escherichia coli cyclophilin A preferably recognizes a cis-proline isomer or a highly distorted form of the trans isomer.
J.Mol.Biol., 256, 1996
|
|
1LOQ
 
 | |
1LOR
 
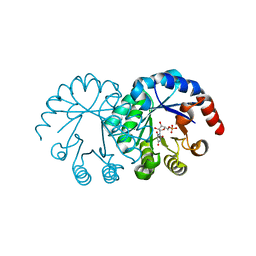 | | crystal structure of orotidine 5'-monophosphate complexed with BMP | | Descriptor: | 6-HYDROXYURIDINE-5'-PHOSPHATE, orotidine monophosphate decarboxylase | | Authors: | Wu, N, Pai, E.F. | | Deposit date: | 2002-05-06 | | Release date: | 2002-08-07 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Crystal structures of inhibitor complexes reveal an alternate binding mode in orotidine-5'-monophosphate decarboxylase.
J.Biol.Chem., 277, 2002
|
|
1LOS
 
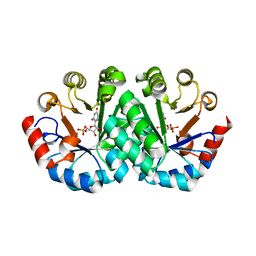 | |
1LOU
 
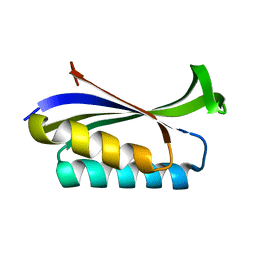 | | RIBOSOMAL PROTEIN S6 | | Descriptor: | RIBOSOMAL PROTEIN S6 | | Authors: | Otzen, D.E, Kristensen, O, Proctor, M, Oliveberg, M. | | Deposit date: | 1998-11-25 | | Release date: | 1998-11-30 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Structural changes in the transition state of protein folding: alternative interpretations of curved chevron plots.
Biochemistry, 38, 1999
|
|
1LOV
 
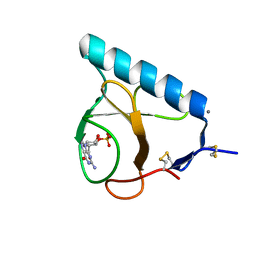 | | X-ray structure of the E58A mutant of Ribonuclease T1 complexed with 3'-guanosine monophosphate | | Descriptor: | CALCIUM ION, GUANOSINE-3'-MONOPHOSPHATE, Guanyl-specific ribonuclease T1 | | Authors: | Mignon, P, Steyaert, J, Loris, R, Geerlings, P, Loverix, S. | | Deposit date: | 2002-05-07 | | Release date: | 2002-08-21 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | A nucleophile activation dyad in ribonucleases. A combined X-ray crystallographic/ab initio quantum chemical study
J.Biol.Chem., 277, 2002
|
|
1LOW
 
 | | X-ray structure of the H40A mutant of Ribonuclease T1 complexed with 3'-guanosine monophosphate | | Descriptor: | CALCIUM ION, GUANOSINE-3'-MONOPHOSPHATE, Guanyl-specific ribonuclease T1 | | Authors: | Mignon, P, Steyaert, J, Loris, R, Geerlings, P, Loverix, S. | | Deposit date: | 2002-05-07 | | Release date: | 2002-08-21 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | A nucleophile activation dyad in ribonucleases. A combined X-ray crystallographic/ab initio quantum chemical study
J.Biol.Chem., 277, 2002
|
|
1LOX
 
 | | RABBIT RETICULOCYTE 15-LIPOXYGENASE | | Descriptor: | (2E)-3-(2-OCT-1-YN-1-YLPHENYL)ACRYLIC ACID, 15-LIPOXYGENASE, FE (II) ION | | Authors: | Gillmor, S.A, Villasenor, A, Fletterick, R.J, Sigal, E, Browner, M.F. | | Deposit date: | 1997-10-06 | | Release date: | 1998-11-04 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | The structure of mammalian 15-lipoxygenase reveals similarity to the lipases and the determinants of substrate specificity.
Nat.Struct.Biol., 4, 1997
|
|
1LOY
 
 | | X-ray structure of the H40A/E58A mutant of Ribonuclease T1 complexed with 3'-guanosine monophosphate | | Descriptor: | CALCIUM ION, GUANOSINE-3'-MONOPHOSPHATE, Guanyl-specific ribonuclease T1 | | Authors: | Mignon, P, Steyaert, J, Loris, R, Geerlings, P, Loverix, S. | | Deposit date: | 2002-05-07 | | Release date: | 2002-08-21 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | A nucleophile activation dyad in ribonucleases. A combined X-ray crystallographic/ab initio quantum chemical study
J.Biol.Chem., 277, 2002
|
|
1LOZ
 
 | |
1LP1
 
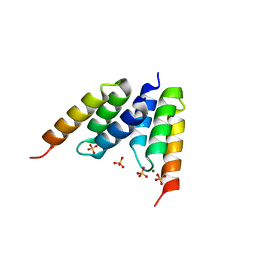 | | Protein Z in complex with an in vitro selected affibody | | Descriptor: | Affibody binding protein Z, Immunoglobulin G binding protein A, MAGNESIUM ION, ... | | Authors: | Hogbom, M, Eklund, M, Nygren, P.A, Nordlund, P. | | Deposit date: | 2002-05-07 | | Release date: | 2003-03-18 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structural basis for recognition by an in vitro evolved affibody.
Proc.Natl.Acad.Sci.USA, 100, 2003
|
|
1LP3
 
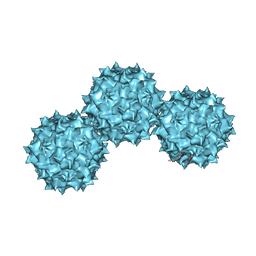 | | The Atomic Structure of Adeno-Associated Virus (AAV-2), a Vector for Human Gene Therapy | | Descriptor: | AAV-2 capsid protein | | Authors: | Xie, Q, Bu, W, Bhatia, S, Hare, J, Somasundaram, T, Azzi, A, Chapman, M.S. | | Deposit date: | 2002-05-07 | | Release date: | 2002-08-07 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | The atomic structure of adeno-associated virus (AAV-2), a vector for human gene therapy.
Proc.Natl.Acad.Sci.USA, 99, 2002
|
|
1LP4
 
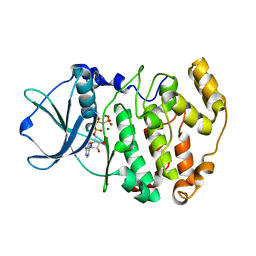 | | Crystal structure of a binary complex of the catalytic subunit of protein kinase CK2 with Mg-AMPPNP | | Descriptor: | MAGNESIUM ION, PHOSPHOAMINOPHOSPHONIC ACID-ADENYLATE ESTER, Protein kinase CK2 | | Authors: | Niefind, K, Puetter, M, Guerra, B, Issinger, O.-G, Schomburg, D. | | Deposit date: | 2002-05-07 | | Release date: | 2002-05-29 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.86 Å) | | Cite: | Inclining the purine base binding plane in protein kinase CK2 by exchanging the flanking side-chains generates a preference for ATP as a cosubstrate.
J.Mol.Biol., 347, 2005
|
|
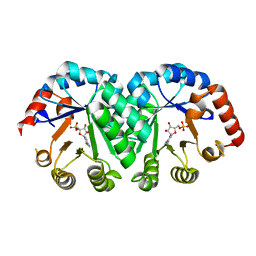
1LP6
 
 | |
1LP9
 
 | | Xenoreactive complex AHIII 12.2 TCR bound to p1049/HLA-A2.1 | | Descriptor: | Beta-2-microglobulin, HLA class I histocompatibility antigen, A-2 alpha chain, ... | | Authors: | Buslepp, J, Wang, H, Biddison, W.E, Appella, E, Collins, E.J. | | Deposit date: | 2002-05-07 | | Release date: | 2003-11-11 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | A correlation between TCR Valpha docking on MHC and CD8 dependence: implications for T cell selection.
Immunity, 19, 2003
|
|
1LPF
 
 | |
1LPG
 
 | | CRYSTAL STRUCTURE OF FXA IN COMPLEX WITH 79. | | Descriptor: | Blood coagulation factor Xa, CALCIUM ION, [4-({[5-BENZYLOXY-1-(3-CARBAMIMIDOYL-BENZYL)-1H-INDOLE-2-CARBONYL]-AMINO}-METHYL)-PHENYL]-TRIMETHYL-AMMONIUM | | Authors: | Schreuder, H.A, Brachvogel, V, Liesum, A. | | Deposit date: | 2002-05-08 | | Release date: | 2003-05-08 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Design and Quantitative Structure-Activity relationship of 3-amidinobenzyl-1H-indole-2-carboxamides as potent, nonchiral, and selective inhibitors of blood coagulation factor Xa.
J.Med.Chem., 45, 2002
|
|
1LPI
 
 | | HEW LYSOZYME: TRP...NA CATION-PI INTERACTION | | Descriptor: | LYSOZYME, SODIUM ION | | Authors: | Wouters, J. | | Deposit date: | 1998-04-15 | | Release date: | 1998-06-17 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Cation-pi (Na+-Trp) interactions in the crystal structure of tetragonal lysozyme.
Protein Sci., 7, 1998
|
|
