1JEC
 
 | |
1JED
 
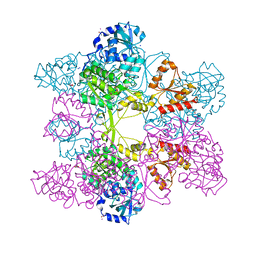 | | Crystal Structure of ATP Sulfurylase in complex with ADP | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, ACETIC ACID, ADENOSINE-5'-DIPHOSPHATE, ... | | Authors: | Ullrich, T.C, Huber, R. | | Deposit date: | 2001-06-17 | | Release date: | 2001-11-14 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2.95 Å) | | Cite: | The complex structures of ATP sulfurylase with thiosulfate, ADP and chlorate reveal new insights in inhibitory effects and the catalytic cycle.
J.Mol.Biol., 313, 2001
|
|
1JEE
 
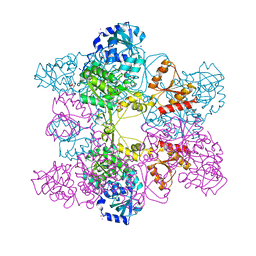 | | Crystal Structure of ATP Sulfurylase in complex with chlorate | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, ACETIC ACID, ADENOSINE-5'-PHOSPHOSULFATE, ... | | Authors: | Ullrich, T.C, Huber, R. | | Deposit date: | 2001-06-17 | | Release date: | 2001-11-14 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | The complex structures of ATP sulfurylase with thiosulfate, ADP and chlorate reveal new insights in inhibitory effects and the catalytic cycle.
J.Mol.Biol., 313, 2001
|
|
1JEF
 
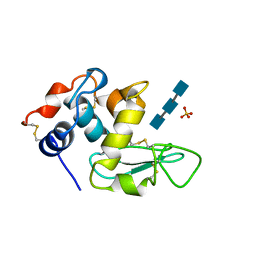 | | TURKEY LYSOZYME COMPLEX WITH (GLCNAC)3 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, LYSOZYME, SULFATE ION | | Authors: | Harata, K, Muraki, M. | | Deposit date: | 1997-04-23 | | Release date: | 1997-10-15 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.77 Å) | | Cite: | X-ray structure of turkey-egg lysozyme complex with tri-N-acetylchitotriose. Lack of binding ability at subsite A.
Acta Crystallogr.,Sect.D, 53, 1997
|
|
1JEJ
 
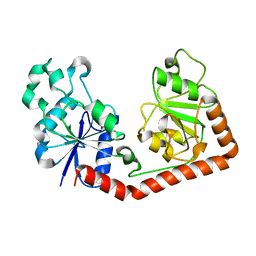 | | T4 phage apo BGT | | Descriptor: | DNA BETA-GLUCOSYLTRANSFERASE | | Authors: | Morera, S, Lariviere, L, Kurzeck, J, Aschke-Sonnenborn, U, Freemont, P.S, Janin, J, Ruger, W. | | Deposit date: | 2001-06-18 | | Release date: | 2001-08-15 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | High resolution crystal structures of T4 phage beta-glucosyltransferase: induced fit and effect of substrate and metal binding.
J.Mol.Biol., 311, 2001
|
|
1JEO
 
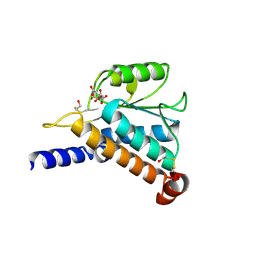 | | Crystal Structure of the Hypothetical Protein MJ1247 from Methanococcus jannaschii at 2.0 A Resolution Infers a Molecular Function of 3-Hexulose-6-Phosphate isomerase. | | Descriptor: | CITRIC ACID, HYPOTHETICAL PROTEIN MJ1247 | | Authors: | Martinez-Cruz, L.A, Dreyer, M.K, Boisvert, D.C, Yokota, H, Martinez-Chantar, M.L, Kim, R, Kim, S.H, Berkeley Structural Genomics Center (BSGC) | | Deposit date: | 2001-06-18 | | Release date: | 2002-02-20 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal structure of MJ1247 protein from M. jannaschii at 2.0 A resolution infers a molecular function of 3-hexulose-6-phosphate isomerase.
Structure, 10, 2002
|
|
1JEP
 
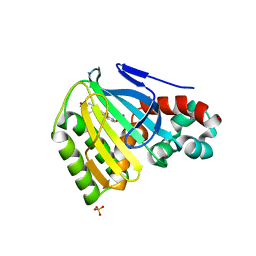 | | Chalcone Isomerase Complexed with 4'-hydroxyflavanone | | Descriptor: | 2-(4-HYDROXY-PHENYL)-CHROMAN-4-ONE, CHALCONE--FLAVONONE ISOMERASE 1, SULFATE ION | | Authors: | Jez, J.M, Noel, J.P. | | Deposit date: | 2001-06-18 | | Release date: | 2001-12-12 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Reaction mechanism of chalcone isomerase. pH dependence, diffusion control, and product binding differences.
J.Biol.Chem., 277, 2002
|
|
1JEQ
 
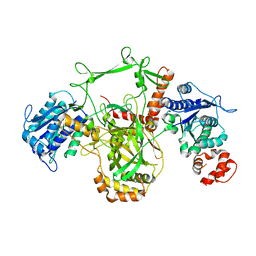 | |
1JER
 
 | | CUCUMBER STELLACYANIN, CU2+, PH 7.0 | | Descriptor: | COPPER (II) ION, CUCUMBER STELLACYANIN | | Authors: | Hart, P.J, Nersissian, A.M, Herrmann, R.G, Nalbandyan, R.M, Valentine, J.S, Eisenberg, D. | | Deposit date: | 1996-08-21 | | Release date: | 1997-02-12 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | A missing link in cupredoxins: crystal structure of cucumber stellacyanin at 1.6 A resolution.
Protein Sci., 5, 1996
|
|
1JES
 
 | | Crystal Structure of a Copper-Mediated Base Pair in DNA | | Descriptor: | 5'-D(*CP*GP*CP*GP*(DPY)P*AP*TP*(DRP)P*CP*GP*CP*G)-3', COPPER (II) ION | | Authors: | Atwell, S, Meggers, E, Spraggon, G, Schultz, P.G. | | Deposit date: | 2001-06-18 | | Release date: | 2001-11-23 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Structure of a Copper-Mediated Base Pair in DNA
J.Am.Chem.Soc., 123, 2001
|
|
1JF1
 
 | | Crystal structure of HLA-A2*0201 in complex with a decameric altered peptide ligand from the MART-1/Melan-A | | Descriptor: | HLA CLASS I HISTOCOMPATIBILITY ANTIGEN, A-2 ALPHA CHAIN, ZINC ION, ... | | Authors: | Sliz, P, Michielin, O, Cerottini, J.C, Luescher, I, Romero, P, Karplus, M, Wiley, D.C. | | Deposit date: | 2001-06-19 | | Release date: | 2001-09-14 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Crystal structures of two closely related but antigenically distinct HLA-A2/melanocyte-melanoma tumor-antigen peptide complexes.
J.Immunol., 167, 2001
|
|
1JF7
 
 | | HUMAN PTP1B CATALYTIC DOMAIN COMPLEXED WITH PNU177836 | | Descriptor: | 5-(2-{2-[(TERT-BUTOXY-HYDROXY-METHYL)-AMINO]-1-HYDROXY-3-PHENYL-PROPYLAMINO}-3-HYDROXY-3-PENTYLAMINO-PROPYL)-2-CARBOXYMETHOXY-BENZOIC ACID, PROTEIN-TYROSINE PHOSPHATASE 1B | | Authors: | Larsen, S.D, Barf, T, Liljebris, C, May, P.D, Ogg, D, O'Sullivan, T.J, Palazuk, B.J, Schostarez, H.J, Stevens, F.C, Bleasdale, J.E. | | Deposit date: | 2001-06-20 | | Release date: | 2002-02-13 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Synthesis and biological activity of a novel class of small molecular weight peptidomimetic competitive inhibitors of protein tyrosine phosphatase 1B.
J.Med.Chem., 45, 2002
|
|
1JF8
 
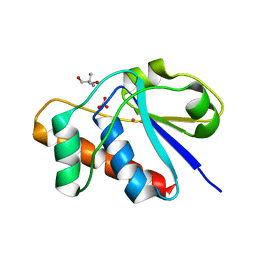 | | X-ray structure of reduced C10S, C15A arsenate reductase from pI258 | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, BICARBONATE ION, POTASSIUM ION, ... | | Authors: | Zegers, I, Martins, J.C, Willem, R, Wyns, L, Messens, J. | | Deposit date: | 2001-06-20 | | Release date: | 2001-10-03 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.12 Å) | | Cite: | Arsenate reductase from S. aureus plasmid pI258 is a phosphatase drafted for redox duty.
Nat.Struct.Biol., 8, 2001
|
|
1JF9
 
 | |
1JFA
 
 | |
1JFG
 
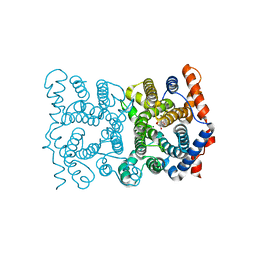 | | TRICHODIENE SYNTHASE FROM FUSARIUM SPOROTRICHIOIDES COMPLEXED WITH DIPHOSPHATE | | Descriptor: | GLYCEROL, MAGNESIUM ION, PYROPHOSPHATE 2-, ... | | Authors: | Rynkiewicz, M.J, Cane, D.E, Christianson, D.W. | | Deposit date: | 2001-06-20 | | Release date: | 2001-11-30 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structure of trichodiene synthase from Fusarium sporotrichioides provides mechanistic inferences on the terpene cyclization cascade.
Proc.Natl.Acad.Sci.USA, 98, 2001
|
|
1JFI
 
 | | Crystal Structure of the NC2-TBP-DNA Ternary Complex | | Descriptor: | 5'-D(*G*GP*AP*GP*CP*CP*CP*TP*TP*TP*TP*AP*TP*AP*GP*CP*CP*AP*A)-3', 5'-D(*TP*TP*GP*GP*CP*TP*AP*TP*AP*AP*AP*AP*GP*GP*GP*CP*TP*CP*C)-3', TATA-BOX-BINDING PROTEIN (TBP), ... | | Authors: | Kamada, K, Shu, F, Chen, H, Malik, S, Stelzer, G, Roeder, R.G, Meisterernst, M, Burley, S.K, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2001-06-20 | | Release date: | 2001-07-11 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2.62 Å) | | Cite: | Crystal structure of negative cofactor 2 recognizing the TBP-DNA transcription complex.
Cell(Cambridge,Mass.), 106, 2001
|
|
1JFU
 
 | | CRYSTAL STRUCTURE OF THE SOLUBLE DOMAIN OF TLPA FROM BRADYRHIZOBIUM JAPONICUM | | Descriptor: | THIOL:DISULFIDE INTERCHANGE PROTEIN TLPA | | Authors: | Capitani, G, Rossmann, R, Sargent, D.F, Gruetter, M.G, Richmond, T.J, Hennecke, H. | | Deposit date: | 2001-06-22 | | Release date: | 2001-09-19 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Structure of the soluble domain of a membrane-anchored thioredoxin-like protein from Bradyrhizobium japonicum reveals unusual properties.
J.Mol.Biol., 311, 2001
|
|
1JFV
 
 | | X-Ray Structure of oxidised C10S, C15A arsenate reductase from pI258 | | Descriptor: | PERCHLORATE ION, POTASSIUM ION, arsenate reductase | | Authors: | Zegers, I, Martins, J.C, Willem, R, Wyns, L, Messens, J. | | Deposit date: | 2001-06-22 | | Release date: | 2001-10-03 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Arsenate reductase from S. aureus plasmid pI258 is a phosphatase drafted for redox duty.
Nat.Struct.Biol., 8, 2001
|
|
1JFX
 
 | | Crystal structure of the bacterial lysozyme from Streptomyces coelicolor at 1.65 A resolution | | Descriptor: | 1,4-beta-N-Acetylmuramidase M1, CHLORIDE ION | | Authors: | Rau, A, Hogg, T, Marquardt, R, Hilgenfeld, R. | | Deposit date: | 2001-06-22 | | Release date: | 2001-09-05 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | A new lysozyme fold. Crystal structure of the muramidase from Streptomyces coelicolor at 1.65 A resolution.
J.Biol.Chem., 276, 2001
|
|
1JFZ
 
 | |
1JG1
 
 | | Crystal Structure of L-isoaspartyl (D-aspartyl) O-methyltransferase with S-ADENOSYL-L-HOMOCYSTEINE | | Descriptor: | S-ADENOSYL-L-HOMOCYSTEINE, protein-L-isoaspartate O-methyltransferase | | Authors: | Griffith, S.C, Sawaya, M.R, Boutz, D, Thapar, N, Katz, J, Clarke, S, Yeates, T.O. | | Deposit date: | 2001-06-22 | | Release date: | 2001-11-16 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.2 Å) | | Cite: | Crystal structure of a protein repair methyltransferase from Pyrococcus furiosus with its L-isoaspartyl peptide substrate.
J.Mol.Biol., 313, 2001
|
|
1JG2
 
 | | Crystal Structure of L-isoaspartyl (D-aspartyl) O-methyltransferase with adenosine | | Descriptor: | ADENOSINE, SODIUM ION, protein-L-isoaspartate O-methyltransferase | | Authors: | Griffith, S.C, Sawaya, M.R, Boutz, D, Thapar, N, Katz, J, Clarke, S, Yeates, T.O. | | Deposit date: | 2001-06-22 | | Release date: | 2001-11-16 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Crystal structure of a protein repair methyltransferase from Pyrococcus furiosus with its L-isoaspartyl peptide substrate.
J.Mol.Biol., 313, 2001
|
|
1JG3
 
 | | Crystal Structure of L-isoaspartyl (D-aspartyl) O-methyltransferase with adenosine & VYP(ISP)HA substrate | | Descriptor: | ADENOSINE, CHLORIDE ION, SODIUM ION, ... | | Authors: | Griffith, S.C, Sawaya, M.R, Boutz, D, Thapar, N, Katz, J, Clarke, S, Yeates, T.O. | | Deposit date: | 2001-06-22 | | Release date: | 2001-11-16 | | Last modified: | 2011-07-27 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Crystal structure of a protein repair methyltransferase from Pyrococcus furiosus with its L-isoaspartyl peptide substrate.
J.Mol.Biol., 313, 2001
|
|
1JG4
 
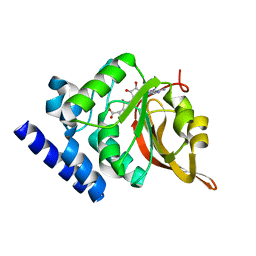 | | Crystal Structure of L-isoaspartyl (D-aspartyl) O-methyltransferase with S-adenosylmethionine | | Descriptor: | S-ADENOSYLMETHIONINE, protein-L-isoaspartate O-methyltransferase | | Authors: | Griffith, S.C, Sawaya, M.R, Boutz, D, Thapar, N, Katz, J, Clarke, S, Yeates, T.O. | | Deposit date: | 2001-06-22 | | Release date: | 2001-11-16 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Crystal structure of a protein repair methyltransferase from Pyrococcus furiosus with its L-isoaspartyl peptide substrate.
J.Mol.Biol., 313, 2001
|
|
