1IDQ
 
 | |
1IDR
 
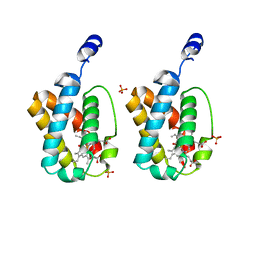 | | CRYSTAL STRUCTURE OF THE TRUNCATED-HEMOGLOBIN-N FROM MYCOBACTERIUM TUBERCULOSIS | | Descriptor: | HEMOGLOBIN HBN, OXYGEN MOLECULE, PHOSPHATE ION, ... | | Authors: | Milani, M, Pesce, A, Ascenzi, P, Guertin, M, Bolognesi, M. | | Deposit date: | 2001-04-05 | | Release date: | 2001-08-22 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Mycobacterium tuberculosis hemoglobin N displays a protein tunnel suited for O2 diffusion to the heme.
EMBO J., 20, 2001
|
|
1IDS
 
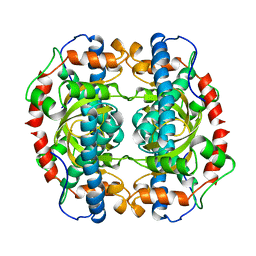 | | X-RAY STRUCTURE ANALYSIS OF THE IRON-DEPENDENT SUPEROXIDE DISMUTASE FROM MYCOBACTERIUM TUBERCULOSIS AT 2.0 ANGSTROMS RESOLUTIONS REVEALS NOVEL DIMER-DIMER INTERACTIONS | | Descriptor: | FE (III) ION, IRON SUPEROXIDE DISMUTASE | | Authors: | Cooper, J.B, Mcintyre, K, Wood, S.P, Zhang, Y, Young, D. | | Deposit date: | 1994-09-29 | | Release date: | 1994-12-20 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | X-ray structure analysis of the iron-dependent superoxide dismutase from Mycobacterium tuberculosis at 2.0 Angstroms resolution reveals novel dimer-dimer interactions.
J.Mol.Biol., 246, 1995
|
|
1IDT
 
 | | STRUCTURAL STUDIES ON A PRODRUG-ACTIVATING SYSTEM-CB1954 AND FMN-DEPENDENT NITROREDUCTASE | | Descriptor: | 5-(AZIRIDIN-1-YL)-2,4-DINITROBENZAMIDE, FLAVIN MONONUCLEOTIDE, MINOR FMN-DEPENDENT NITROREDUCTASE | | Authors: | Johansson, E, Parkinson, G.N, Denny, W.A, Neidle, S. | | Deposit date: | 2001-04-05 | | Release date: | 2003-09-16 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Studies on the Nitroreductase Prodrug-Activating System. Crystal Structures of Complexes with the Inhibitor Dicoumarol and Dinitrobenzamide Prodrugs and of the Enzyme Active Form.
J.Med.Chem., 46, 2003
|
|
1IDU
 
 | |
1IE0
 
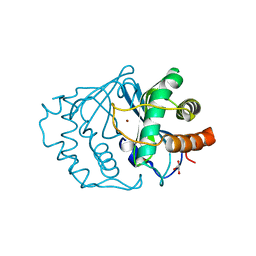 | | CRYSTAL STRUCTURE OF LUXS | | Descriptor: | AUTOINDUCER-2 PRODUCTION PROTEIN LUXS, GLYCEROL, ZINC ION | | Authors: | Hilgers, M.T, Ludwig, M.L. | | Deposit date: | 2001-04-05 | | Release date: | 2001-10-03 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Crystal structure of the quorum-sensing protein LuxS reveals a catalytic metal site.
Proc.Natl.Acad.Sci.USA, 98, 2001
|
|
1IE3
 
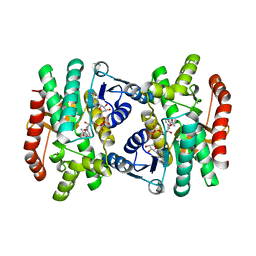 | | CRYSTAL STRUCTURE OF R153C E. COLI MALATE DEHYDROGENASE | | Descriptor: | MALATE DEHYDROGENASE, NICOTINAMIDE-ADENINE-DINUCLEOTIDE, PYRUVIC ACID | | Authors: | Bell, J.K, Yennawar, H.P, Wright, S.K, Thompson, J.R, Viola, R.E, Banaszak, L.J. | | Deposit date: | 2001-04-05 | | Release date: | 2001-09-19 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structural Analyses of a Malate Dehydrogenase with a Variable Active Site
J.Biol.Chem., 276, 2001
|
|
1IE7
 
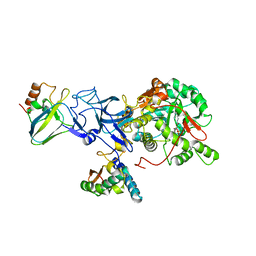 | | PHOSPHATE INHIBITED BACILLUS PASTEURII UREASE CRYSTAL STRUCTURE | | Descriptor: | NICKEL (II) ION, PHOSPHATE ION, UREASE ALPHA SUBUNIT, ... | | Authors: | Benini, S, Rypniewski, W.R, Wilson, K.S, Ciurli, S, Mangani, S. | | Deposit date: | 2001-04-09 | | Release date: | 2001-04-25 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Structure-based rationalization of urease inhibition by phosphate: novel insights into the enzyme mechanism.
J.Biol.Inorg.Chem., 6, 2001
|
|
1IE8
 
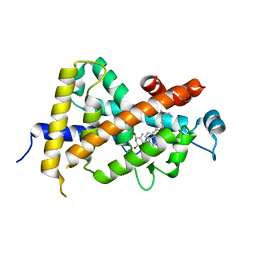 | | Crystal Structure Of The Nuclear Receptor For Vitamin D Ligand Binding Domain Bound to KH1060 | | Descriptor: | 5-(2-{1-[1-(4-ETHYL-4-HYDROXY-HEXYLOXY)-ETHYL]-7A-METHYL-OCTAHYDRO-INDEN-4-YLIDENE}-ETHYLIDENE)-4-METHYLENE-CYCLOHEXANE-1,3-DIOL, VITAMIN D3 RECEPTOR | | Authors: | Tocchini-Valentini, G, Rochel, N, Wurtz, J.M, Mitschler, A, Moras, D. | | Deposit date: | 2001-04-09 | | Release date: | 2001-05-16 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.52 Å) | | Cite: | Crystal structures of the vitamin D receptor complexed to superagonist 20-epi ligands.
Proc.Natl.Acad.Sci.USA, 98, 2001
|
|
1IE9
 
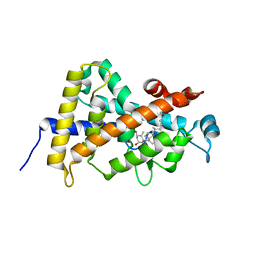 | | Crystal Structure Of The Nuclear Receptor For Vitamin D Ligand Binding Domain Bound to MC1288 | | Descriptor: | 5-{2-[1-(5-HYDROXY-1,5-DIMETHYL-HEXYL)-7A-METHYL-OCTAHYDRO-INDEN-4-YLIDENE]-ETHYLIDENE}-4-METHYLENE-CYCLOHEXANE-1,3-DIOL, VITAMIN D3 RECEPTOR | | Authors: | Tocchini-Valentini, G, Rochel, N, Wurtz, J.M, Mitschler, A, Moras, D. | | Deposit date: | 2001-04-09 | | Release date: | 2001-05-16 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Crystal structures of the vitamin D receptor complexed to superagonist 20-epi ligands.
Proc.Natl.Acad.Sci.USA, 98, 2001
|
|
1IEE
 
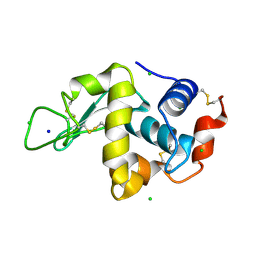 | | STRUCTURE OF TETRAGONAL HEN EGG WHITE LYSOZYME AT 0.94 A FROM CRYSTALS GROWN BY THE COUNTER-DIFFUSION METHOD | | Descriptor: | CHLORIDE ION, LYSOZYME C, SODIUM ION | | Authors: | Sauter, C, Otalora, F, Gavira, J.-A, Vidal, O, Giege, R, Garcia-Ruiz, J.-M. | | Deposit date: | 2001-04-09 | | Release date: | 2001-08-08 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (0.94 Å) | | Cite: | Structure of tetragonal hen egg-white lysozyme at 0.94 A from crystals grown by the counter-diffusion method.
Acta Crystallogr.,Sect.D, 57, 2001
|
|
1IEI
 
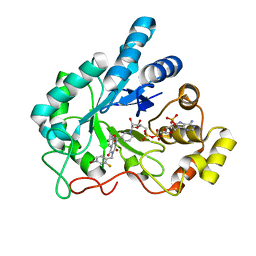 | | CRYSTAL STRUCTURE OF HUMAN ALDOSE REDUCTASE COMPLEXED WITH THE INHIBITOR ZENARESTAT. | | Descriptor: | ALDOSE REDUCTASE, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, [3-(4-BROMO-2-FLUORO-BENZYL)-7-CHLORO-2,4-DIOXO-3,4-DIHYDRO-2H-QUINAZOLIN-1-YL]-ACETIC ACID | | Authors: | Kinoshita, T, Miyake, H, Fujii, T, Takakura, S, Goto, T. | | Deposit date: | 2001-04-09 | | Release date: | 2002-04-10 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | The structure of human recombinant aldose reductase complexed with the potent inhibitor zenarestat.
Acta Crystallogr.,Sect.D, 58, 2002
|
|
1IEL
 
 | | Crystal Structure of AmpC beta-lactamase from E. coli in Complex with Ceftazidime | | Descriptor: | ACYLATED CEFTAZIDIME, PHOSPHATE ION, beta-lactamase | | Authors: | Powers, R.A, Caselli, E, Focia, P.J, Prati, F, Shoichet, B.K. | | Deposit date: | 2001-04-10 | | Release date: | 2001-08-15 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structures of ceftazidime and its transition-state analogue in complex with AmpC beta-lactamase: implications for resistance mutations and inhibitor design.
Biochemistry, 40, 2001
|
|
1IEM
 
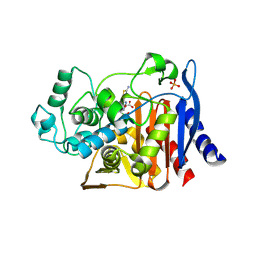 | | Crystal Structure of AmpC beta-lactamase from E. coli in Complex with a Boronic Acid Inhibitor (1, CefB4) | | Descriptor: | PHOSPHATE ION, PINACOL[[2-AMINO-ALPHA-(1-CARBOXY-1-METHYLETHOXYIMINO)-4-THIAZOLEACETYL]AMINO]METHANEBORONATE, beta-lactamase | | Authors: | Powers, R.A, Caselli, E, Focia, P.J, Prati, F, Shoichet, B.K. | | Deposit date: | 2001-04-10 | | Release date: | 2001-08-15 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structures of ceftazidime and its transition-state analogue in complex with AmpC beta-lactamase: implications for resistance mutations and inhibitor design.
Biochemistry, 40, 2001
|
|
1IEP
 
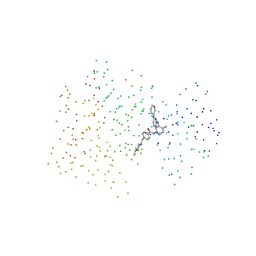 | | CRYSTAL STRUCTURE OF THE C-ABL KINASE DOMAIN IN COMPLEX WITH STI-571. | | Descriptor: | 4-(4-METHYL-PIPERAZIN-1-YLMETHYL)-N-[4-METHYL-3-(4-PYRIDIN-3-YL-PYRIMIDIN-2-YLAMINO)-PHENYL]-BENZAMIDE, CHLORIDE ION, PROTO-ONCOGENE TYROSINE-PROTEIN KINASE ABL | | Authors: | Nagar, B, Bornmann, W, Schindler, T, Clarkson, B, Kuriyan, J. | | Deposit date: | 2001-04-10 | | Release date: | 2001-04-18 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Crystal structures of the kinase domain of c-Abl in complex with the small molecule inhibitors PD173955 and imatinib (STI-571)
Cancer Res., 62, 2002
|
|
1IF2
 
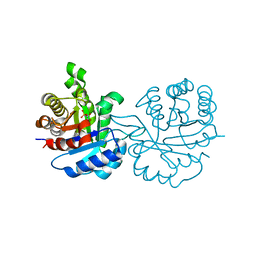 | | X-RAY STRUCTURE OF LEISHMANIA MEXICANA TRIOSEPHOSPHATE ISOMERASE COMPLEXED WITH IPP | | Descriptor: | TRIOSEPHOSPHATE ISOMERASE, [2(FORMYL-HYDROXY-AMINO)-ETHYL]-PHOSPHONIC ACID | | Authors: | Kursula, I, Partanen, S, Lambeir, A.-M, Antonov, D.M, Augustyns, K, Wierenga, R.K. | | Deposit date: | 2001-04-12 | | Release date: | 2001-08-17 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural determinants for ligand binding and catalysis of triosephosphate isomerase.
Eur.J.Biochem., 268, 2001
|
|
1IFH
 
 | |
1IFP
 
 | |
1IFQ
 
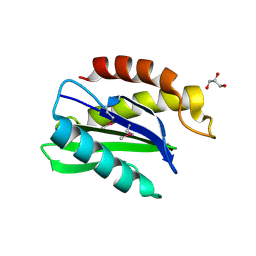 | | Sec22b N-terminal domain | | Descriptor: | GLYCEROL, vesicle trafficking protein Sec22b | | Authors: | Gonzalez Jr, L.C, Weis, W.I, Scheller, R.H. | | Deposit date: | 2001-04-13 | | Release date: | 2001-05-02 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | A novel snare N-terminal domain revealed by the crystal structure of Sec22b.
J.Biol.Chem., 276, 2001
|
|
1IFV
 
 | |
1IFX
 
 | | CRYSTAL STRUCTURE OF NH3-DEPENDENT NAD+ SYNTHETASE FROM BACILLUS SUBTILIS COMPLEXED WITH TWO MOLECULES DEAMIDO-NAD | | Descriptor: | NH(3)-DEPENDENT NAD(+) SYNTHETASE, NICOTINIC ACID ADENINE DINUCLEOTIDE | | Authors: | Devedjiev, Y, Symersky, J, Singh, R, Brouillette, W, Muccio, D, Jedrzejas, M, Brouillette, C, DeLucas, L. | | Deposit date: | 2001-04-13 | | Release date: | 2001-06-06 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Stabilization of active-site loops in NH3-dependent NAD+ synthetase from Bacillus subtilis.
Acta Crystallogr.,Sect.D, 57, 2001
|
|
1IG1
 
 | | 1.8A X-Ray structure of ternary complex of a catalytic domain of death-associated protein kinase with ATP analogue and Mn. | | Descriptor: | MANGANESE (II) ION, PHOSPHOAMINOPHOSPHONIC ACID-ADENYLATE ESTER, death-associated protein kinase | | Authors: | Tereshko, V, Teplova, M, Brunzelle, J, Watterson, D.M, Egli, M. | | Deposit date: | 2001-04-16 | | Release date: | 2002-04-01 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal structures of the catalytic domain of human protein kinase associated with apoptosis and tumor suppression.
Nat.Struct.Biol., 8, 2001
|
|
1IG3
 
 | | Mouse Thiamin Pyrophosphokinase Complexed with Thiamin | | Descriptor: | 3-(4-AMINO-2-METHYL-PYRIMIDIN-5-YLMETHYL)-5-(2-HYDROXY-ETHYL)-4-METHYL-THIAZOL-3-IUM, SULFATE ION, thiamin pyrophosphokinase | | Authors: | Timm, D.E, Liu, J, Baker, L.-J, Harris, R.A. | | Deposit date: | 2001-04-16 | | Release date: | 2001-04-25 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal structure of thiamin pyrophosphokinase.
J.Mol.Biol., 310, 2001
|
|
1IG5
 
 | | BOVINE CALBINDIN D9K BINDING MG2+ | | Descriptor: | MAGNESIUM ION, VITAMIN D-DEPENDENT CALCIUM-BINDING PROTEIN, INTESTINAL | | Authors: | Andersson, E.M, Svensson, L.A. | | Deposit date: | 2001-04-17 | | Release date: | 2001-04-25 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Structural basis for the negative allostery between Ca(2+)- and Mg(2+)-binding in the intracellular Ca(2+)-receptor calbindin D9k.
Protein Sci., 6, 1997
|
|
1IG9
 
 | | Structure of the Replicating Complex of a Pol Alpha Family DNA Polymerase | | Descriptor: | 5'-D(*AP*CP*AP*GP*GP*TP*AP*AP*GP*CP*AP*GP*TP*CP*CP*GP*CP*G)-3', 5'-D(*GP*CP*GP*GP*AP*CP*TP*GP*CP*TP*TP*AP*CP*(DOC))-3', CALCIUM ION, ... | | Authors: | Franklin, M.C, Wang, J, Steitz, T.A. | | Deposit date: | 2001-04-17 | | Release date: | 2001-06-11 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Structure of the Replicating Complex of a Pol Alpha Family DNA Polymerase
Cell(Cambridge,Mass.), 105, 2001
|
|
