100D
 
 | |
101M
 
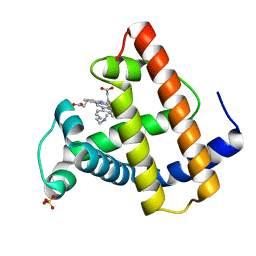 | | SPERM WHALE MYOGLOBIN F46V N-BUTYL ISOCYANIDE AT PH 9.0 | | Descriptor: | MYOGLOBIN, N-BUTYL ISOCYANIDE, PROTOPORPHYRIN IX CONTAINING FE, ... | | Authors: | Smith, R.D, Olson, J.S, Phillips Jr, G.N. | | Deposit date: | 1997-12-13 | | Release date: | 1998-04-08 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.07 Å) | | Cite: | Correlations between Bound N-Alkyl Isocyanide Orientations and Pathways for Ligand Binding in Recombinant Myoglobins
Thesis, Rice, 1999
|
|
102D
 
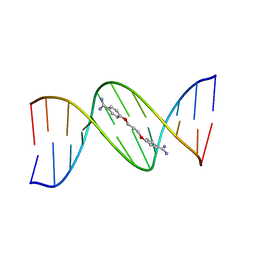 | |
102L
 
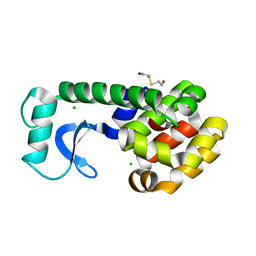 | |
102M
 
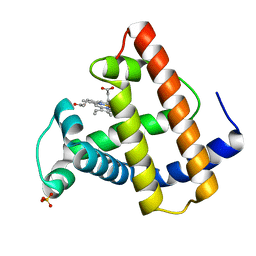 | | SPERM WHALE MYOGLOBIN H64A AQUOMET AT PH 9.0 | | Descriptor: | MYOGLOBIN, PROTOPORPHYRIN IX CONTAINING FE, SULFATE ION | | Authors: | Smith, R.D, Olson, J.S, Phillips Jr, G.N. | | Deposit date: | 1997-12-15 | | Release date: | 1998-04-08 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.84 Å) | | Cite: | Correlations between Bound N-Alkyl Isocyanide Orientations and Pathways for Ligand Binding in Recombinant Myoglobins
Thesis, Rice, 1999
|
|
103L
 
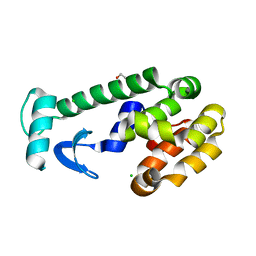 | |
103M
 
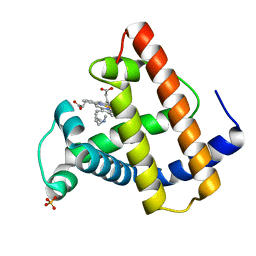 | | SPERM WHALE MYOGLOBIN H64A N-BUTYL ISOCYANIDE AT PH 9.0 | | Descriptor: | MYOGLOBIN, N-BUTYL ISOCYANIDE, PROTOPORPHYRIN IX CONTAINING FE, ... | | Authors: | Smith, R.D, Olson, J.S, Phillips Jr, G.N. | | Deposit date: | 1997-12-16 | | Release date: | 1998-04-08 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.07 Å) | | Cite: | Correlations between Bound N-Alkyl Isocyanide Orientations and Pathways for Ligand Binding in Recombinant Myoglobins
Thesis, Rice, 1999
|
|
104L
 
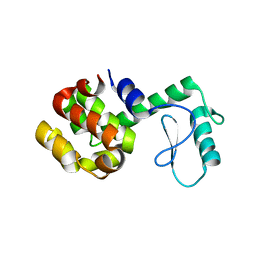 | |
104M
 
 | | SPERM WHALE MYOGLOBIN N-BUTYL ISOCYANIDE AT PH 7.0 | | Descriptor: | MYOGLOBIN, N-BUTYL ISOCYANIDE, PROTOPORPHYRIN IX CONTAINING FE, ... | | Authors: | Smith, R.D, Olson, J.S, Phillips Jr, G.N. | | Deposit date: | 1997-12-18 | | Release date: | 1998-04-08 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.71 Å) | | Cite: | Correlations between Bound N-Alkyl Isocyanide Orientations and Pathways for Ligand Binding in Recombinant Myoglobins
Thesis, Rice, 1999
|
|
105M
 
 | | SPERM WHALE MYOGLOBIN N-BUTYL ISOCYANIDE AT PH 9.0 | | Descriptor: | MYOGLOBIN, N-BUTYL ISOCYANIDE, PROTOPORPHYRIN IX CONTAINING FE, ... | | Authors: | Smith, R.D, Olson, J.S, Phillips Jr, G.N. | | Deposit date: | 1997-12-18 | | Release date: | 1998-04-08 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.02 Å) | | Cite: | Correlations between Bound N-Alkyl Isocyanide Orientations and Pathways for Ligand Binding in Recombinant Myoglobins
Thesis, Rice, 1999
|
|
106M
 
 | | SPERM WHALE MYOGLOBIN V68F ETHYL ISOCYANIDE AT PH 9.0 | | Descriptor: | ETHYL ISOCYANIDE, MYOGLOBIN, PROTOPORPHYRIN IX CONTAINING FE, ... | | Authors: | Smith, R.D, Olson, J.S, Phillips Jr, G.N. | | Deposit date: | 1997-12-21 | | Release date: | 1998-04-08 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.99 Å) | | Cite: | Correlations between Bound N-Alkyl Isocyanide Orientations and Pathways for Ligand Binding in Recombinant Myoglobins
Thesis, Rice, 1999
|
|
107L
 
 | |
107M
 
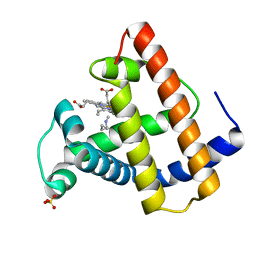 | | SPERM WHALE MYOGLOBIN V68F N-BUTYL ISOCYANIDE AT PH 9.0 | | Descriptor: | MYOGLOBIN, N-BUTYL ISOCYANIDE, PROTOPORPHYRIN IX CONTAINING FE, ... | | Authors: | Smith, R.D, Olson, J.S, Phillips Jr, G.N. | | Deposit date: | 1997-12-22 | | Release date: | 1998-04-08 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.09 Å) | | Cite: | Correlations between Bound N-Alkyl Isocyanide Orientations and Pathways for Ligand Binding in Recombinant Myoglobins
Thesis, Rice, 1999
|
|
108L
 
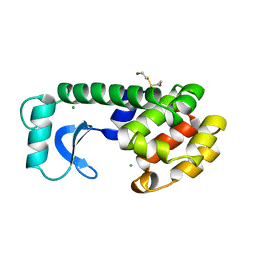 | |
108M
 
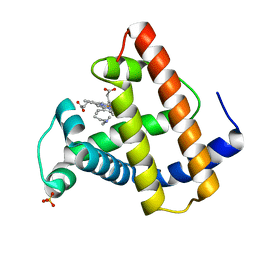 | | SPERM WHALE MYOGLOBIN V68F N-BUTYL ISOCYANIDE AT PH 7.0 | | Descriptor: | MYOGLOBIN, N-BUTYL ISOCYANIDE, PROTOPORPHYRIN IX CONTAINING FE, ... | | Authors: | Smith, R.D, Olson, J.S, Phillips Jr, G.N. | | Deposit date: | 1997-12-23 | | Release date: | 1998-05-20 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.67 Å) | | Cite: | Correlations between Bound N-Alkyl Isocyanide Orientations and Pathways for Ligand Binding in Recombinant Myoglobins
Thesis, Rice, 1999
|
|
109D
 
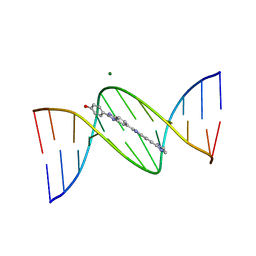 | | VARIABILITY IN DNA MINOR GROOVE WIDTH RECOGNISED BY LIGAND BINDING: THE CRYSTAL STRUCTURE OF A BIS-BENZIMIDAZOLE COMPOUND BOUND TO THE DNA DUPLEX D(CGCGAATTCGCG)2 | | Descriptor: | 5-(2-IMIDAZOLINYL)-2-[2-(4-HYDROXYPHENYL)-5-BENZIMIDAZOLYL]BENZIMIDAZOLE, DNA (5'-D(*CP*GP*CP*GP*AP*AP*TP*TP*CP*GP*CP*G)-3'), MAGNESIUM ION | | Authors: | Czarny, A, Boykin, D.W, Wood, A.A, Nunn, C.M, Neidle, S, Zhao, M, Wilson, W.D. | | Deposit date: | 1995-02-15 | | Release date: | 1995-05-08 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Variability in DNA minor groove width recognised by ligand binding: the crystal structure of a bis-benzimidazole compound bound to the DNA duplex d(CGCGAATTCGCG)2.
Nucleic Acids Res., 23, 1995
|
|
109L
 
 | |
109M
 
 | | SPERM WHALE MYOGLOBIN D122N ETHYL ISOCYANIDE AT PH 9.0 | | Descriptor: | ETHYL ISOCYANIDE, MYOGLOBIN, PROTOPORPHYRIN IX CONTAINING FE, ... | | Authors: | Smith, R.D, Olson, J.S, Phillips Jr, G.N. | | Deposit date: | 1997-12-22 | | Release date: | 1998-04-08 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.83 Å) | | Cite: | Correlations between Bound N-Alkyl Isocyanide Orientations and Pathways for Ligand Binding in Recombinant Myoglobins
Thesis, Rice, 1999
|
|
10AF
 
 | |
10BL
 
 | |
10BM
 
 | |
10GS
 
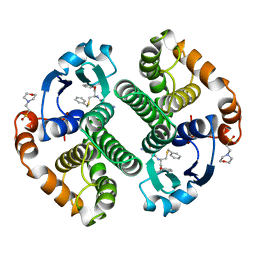 | | HUMAN GLUTATHIONE S-TRANSFERASE P1-1, COMPLEX WITH TER117 | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, GLUTATHIONE S-TRANSFERASE P1-1, L-gamma-glutamyl-S-benzyl-N-[(S)-carboxy(phenyl)methyl]-L-cysteinamide | | Authors: | Oakley, A, Parker, M. | | Deposit date: | 1997-08-14 | | Release date: | 1998-09-16 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | The structures of human glutathione transferase P1-1 in complex with glutathione and various inhibitors at high resolution.
J.Mol.Biol., 274, 1997
|
|
10JU
 
 | |
10KE
 
 | |
10LG
 
 | |
