9GRK
 
 | | Cdc42 Binding peptide (W14A) | | Descriptor: | PRO-SER-ILE-CYS-HIS-VAL-HIS-ARG-PRO-ASP-TRP-PRO-CYS-ALA-TYR-ARG | | Authors: | Mott, H.R, Murphy, N.P, Owen, D. | | Deposit date: | 2024-09-11 | | Release date: | 2026-02-11 | | Method: | SOLUTION NMR | | Cite: | Cyclized Peptide Inhibitors of the Small G Protein Cdc42 Mimic Binding of Effector Proteins.
Biochemistry, 2026
|
|
9M8W
 
 | |
9M8X
 
 | |
9M9G
 
 | |
9M9H
 
 | |
9VA0
 
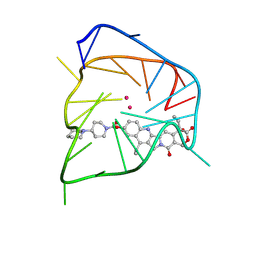 | | Solution NMR structures of hTERT DNA G-quadruplexes (G4s) in complex with small molecule CPT-11 | | Descriptor: | (4S)-4,11-DIETHYL-4-HYDROXY-3,14-DIOXO-3,4,12,14-TETRAHYDRO-1H-PYRANO[3',4':6,7]INDOLIZINO[1,2-B]QUINOLIN-9-YL 1,4'-BIPIPERIDINE-1'-CARBOXYLATE, DNA (5'-D(*AP*AP*TP*GP*GP*GP*AP*GP*GP*GP*TP*CP*TP*GP*GP*GP*AP*GP*GP*GP*CP*C)-3'), POTASSIUM ION | | Authors: | Zeng, L. | | Deposit date: | 2025-06-02 | | Release date: | 2026-02-11 | | Method: | SOLUTION NMR | | Cite: | Dual Targeting of Topoisomerase I and DNA G-quadruplexes Enhances Senescence Response and Chemosensitivity in Colorectal Cancer Cells
To Be Published
|
|
9VA3
 
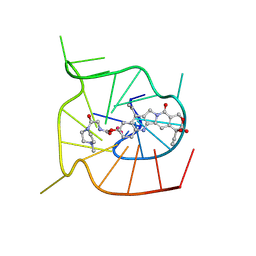 | | Solution NMR structures of hTERT DNA G-quadruplexes (G4s) in complex with small molecule ZBH-01 | | Descriptor: | (S)-4,11-diethyl-4-hydroxy-3,14-dioxo-3,4,12,14-tetrahydro-1H-pyrano[3',4':6,7]indolizino[1,2-b]quinolin-9-yl (2-(4-methylpiperazin-1-yl)-2-oxoethyl)carbamate, DNA (5'-D(*AP*AP*TP*GP*GP*GP*AP*GP*GP*GP*TP*CP*TP*GP*GP*GP*AP*GP*GP*GP*CP*C)-3'), POTASSIUM ION | | Authors: | Zeng, L. | | Deposit date: | 2025-06-02 | | Release date: | 2026-02-11 | | Method: | SOLUTION NMR | | Cite: | Dual Targeting of Topoisomerase I and DNA G-quadruplexes Enhances Senescence Response and Chemosensitivity in Colorectal Cancer Cells
To Be Published
|
|
9VQ1
 
 | |
9X6X
 
 | | Hsp90a N-terminal domain | | Descriptor: | Heat shock protein HSP 90-alpha | | Authors: | Wan, C.J. | | Deposit date: | 2025-10-16 | | Release date: | 2026-02-11 | | Method: | SOLUTION NMR | | Cite: | Phosphorylation regulates Hsp90's ATPase activity
To Be Published
|
|
9X70
 
 | | Hsp90a T36E N-terminal domain | | Descriptor: | Heat shock protein HSP 90-alpha | | Authors: | Wan, C.J. | | Deposit date: | 2025-10-16 | | Release date: | 2026-02-11 | | Method: | SOLUTION NMR | | Cite: | Phosphorylation regulates Hsp90's ATPase activity
To Be Published
|
|
9X71
 
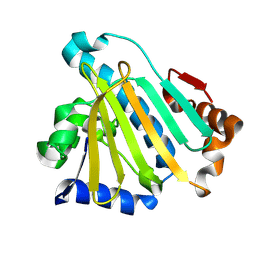 | | the closed-form Hsp90a NTD | | Descriptor: | Heat shock protein HSP 90-alpha | | Authors: | Wan, C.J. | | Deposit date: | 2025-10-16 | | Release date: | 2026-02-11 | | Method: | SOLUTION NMR | | Cite: | Phosphorylation regulates Hsp90's ATPase activity
To Be Published
|
|
9X72
 
 | |
9X73
 
 | | Hsp90a T115E N-terminal domain | | Descriptor: | Heat shock protein HSP 90-alpha | | Authors: | Wan, C.J. | | Deposit date: | 2025-10-16 | | Release date: | 2026-02-11 | | Method: | SOLUTION NMR | | Cite: | Phosphorylation regulates Hsp90's ATPase activity
To Be Published
|
|
