5LWJ
 
 | | Solution NMR structure of the GTP binding Class II RNA aptamer-ligand-complex containing a protonated adenine nucleotide with a highly shifted pKa. | | Descriptor: | GTP Class II RNA (34-MER), GUANOSINE-5'-TRIPHOSPHATE | | Authors: | Wolter, A.C, Weickhmann, A.K, Nasiri, A.H, Hantke, K, Ohlenschlaeger, O, Wunderlich, C.H, Kreutz, C, Duchardt-Ferner, E, Woehnert, J. | | Deposit date: | 2016-09-17 | | Release date: | 2016-12-14 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | A Stably Protonated Adenine Nucleotide with a Highly Shifted pKa Value Stabilizes the Tertiary Structure of a GTP-Binding RNA Aptamer.
Angew. Chem. Int. Ed. Engl., 56, 2017
|
|
5LXJ
 
 | |
5LXK
 
 | | NMR structure of the C-terminal domain of the Bacteriophage T5 decoration protein pb10. | | Descriptor: | Decoration protein | | Authors: | Vernhes, E, Gilquin, B, Cuniasse, P, Boulanger, P, Zinn-Justin, S. | | Deposit date: | 2016-09-22 | | Release date: | 2017-08-02 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | High affinity anchoring of the decoration protein pb10 onto the bacteriophage T5 capsid.
Sci Rep, 7, 2017
|
|
5LXL
 
 | | NMR structure of the N-terminal domain of the Bacteriophage T5 decoration protein pb10 | | Descriptor: | Decoration protein | | Authors: | Vernhes, E, Gilquin, B, Cuniasse, P, Boulanger, P, Zinn-Justin, S. | | Deposit date: | 2016-09-22 | | Release date: | 2017-04-19 | | Last modified: | 2024-06-19 | | Method: | SOLUTION NMR | | Cite: | High affinity anchoring of the decoration protein pb10 onto the bacteriophage T5 capsid.
Sci Rep, 7, 2017
|
|
5M0A
 
 | |
5M1G
 
 | |
5M1H
 
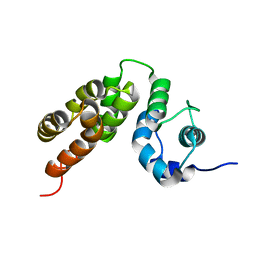 | |
5M1L
 
 | |
5M1U
 
 | |
5M1W
 
 | | Structure of a stable G-hairpin | | Descriptor: | DNA (5'-D(*GP*TP*GP*TP*GP*GP*GP*TP*GP*TP*G)-3') | | Authors: | Gajarsky, M, Zivkovic, M.L, Stadlbauer, P, Pagano, B, Fiala, R, Amato, J, Tomaska, L, Sponer, J, Plavec, J, Trantirek, L. | | Deposit date: | 2016-10-11 | | Release date: | 2017-03-01 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Structure of a Stable G-Hairpin.
J. Am. Chem. Soc., 139, 2017
|
|
5M2L
 
 | |
5M4T
 
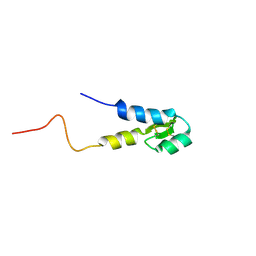 | |
5M4W
 
 | |
5M8I
 
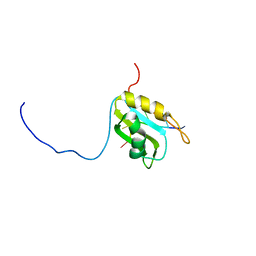 | |
5M9D
 
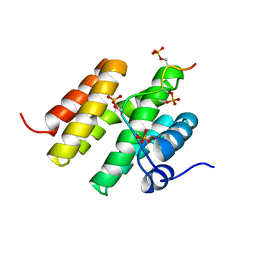 | |
5M9U
 
 | |
5M9Y
 
 | | NMR solution structure of Harzianin HK-VI in DPC micelles | | Descriptor: | Harzianin HK-VI | | Authors: | Kara, S, Zamora-Carreras, H, Afonin, S, Grage, S.L, Ulrich, A.S, Jimenez, M.A. | | Deposit date: | 2016-11-02 | | Release date: | 2018-02-28 | | Last modified: | 2023-11-15 | | Method: | SOLUTION NMR | | Cite: | 11-mer peptaibol Harzianin VI: conformational and biological analysis
To Be Published
|
|
5M9Z
 
 | | Second zinc-binding domain from yeast Pcf11 | | Descriptor: | Protein PCF11, ZINC ION | | Authors: | Mackereth, C. | | Deposit date: | 2016-11-02 | | Release date: | 2017-08-02 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Distinct roles of Pcf11 zinc-binding domains in pre-mRNA 3'-end processing.
Nucleic Acids Res., 45, 2017
|
|
5MBR
 
 | |
5MCR
 
 | | Quadruplex with flipped tetrad formed by an artificial sequence | | Descriptor: | Artificial quadruplex with propeller, diagonal and lateral loop | | Authors: | Dickerhoff, J, Haase, L, Langel, W, Weisz, K. | | Deposit date: | 2016-11-10 | | Release date: | 2017-04-19 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Tracing Effects of Fluorine Substitutions on G-Quadruplex Conformational Changes.
ACS Chem. Biol., 12, 2017
|
|
5MCS
 
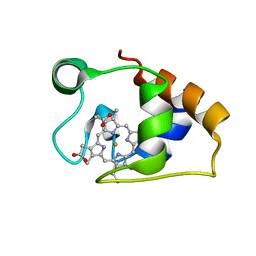 | | Solution structure and dynamics of the outer membrane cytochrome OmcF from Geobacter sulfurreducens | | Descriptor: | HEME C, Lipoprotein cytochrome c, 1 heme-binding site | | Authors: | Dantas, J.M, Silva, M.A, Morgado, L, Pantoja-Uceda, D, Turner, D.L, Bruix, M, Salgueiro, C.A. | | Deposit date: | 2016-11-10 | | Release date: | 2017-04-12 | | Last modified: | 2024-10-23 | | Method: | SOLUTION NMR | | Cite: | Solution structure and dynamics of the outer membrane cytochrome OmcF from Geobacter sulfurreducens.
Biochim. Biophys. Acta, 1858, 2017
|
|
5MF3
 
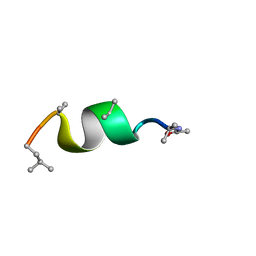 | | NMR solution structure of Harzianin HK-VI in SDS micelles | | Descriptor: | Harzianin HK-VI | | Authors: | Kara, S, Zamora-Carreras, H, Afonin, S, Grage, S.L, Ulrich, A.S, Jimenez, M.A. | | Deposit date: | 2016-11-17 | | Release date: | 2018-06-13 | | Last modified: | 2023-11-15 | | Method: | SOLUTION NMR | | Cite: | 11-mer peptaibol Harzianin HK-VI: conformational and biological analysis
To Be Published
|
|
5MF8
 
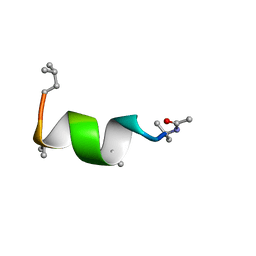 | | NMR solution structure of Harzianin HK-VI in trifluoroethanol | | Descriptor: | Harzianin HK-VI | | Authors: | Kara, S, Zamora-Carreras, H, Afonin, S, Grage, S.L, Ulrich, A.S, Jimenez, M.A. | | Deposit date: | 2016-11-17 | | Release date: | 2018-06-13 | | Last modified: | 2023-11-15 | | Method: | SOLUTION NMR | | Cite: | 11-mer peptaibol Harzianin HK-VI: conformational and biological analysis
To Be Published
|
|
5MF9
 
 | | Solution structure of the RBM5 OCRE domain in complex with polyproline SmN peptide. | | Descriptor: | RNA-binding protein 5, Survival motor neuron protein | | Authors: | Mourao, A, Sattler, M, Bonnal, S, Komal, S, Warner, L, Bordonne, R, Valcarcel, J. | | Deposit date: | 2016-11-17 | | Release date: | 2016-12-07 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Structural basis for the recognition ofspliceosomal SmN B B proteins by theRBM5 OCRE domain in splicing regulation
Elife, 5, 2016
|
|
5MFY
 
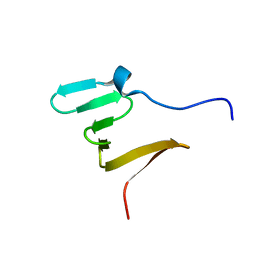 | | RBM5 OCRE domain | | Descriptor: | RNA-binding protein 5 | | Authors: | Warner, L.R, Mourao, A, Soni, K, Sattler, M. | | Deposit date: | 2016-11-19 | | Release date: | 2016-12-07 | | Last modified: | 2024-06-19 | | Method: | SOLUTION NMR | | Cite: | Structural basis for the recognition of spliceosomal SmN/B/B' proteins by the RBM5 OCRE domain in splicing regulation.
Elife, 5, 2016
|
|
