7BQB
 
 | | Solution NMR structure of NF6; de novo designed protein with a novel fold | | Descriptor: | NF6 | | Authors: | Kobayashi, N, Nagashima, T, Minami, S, Koga, R, Chikenji, G, Koga, N. | | Deposit date: | 2020-03-24 | | Release date: | 2021-03-24 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Exploration of novel alpha-beta-protein folds through de novo design
Nat.Struct.Mol.Biol., 2023
|
|
7BQC
 
 | | Solution NMR structure of NF4; de novo designed protein with a novel fold | | Descriptor: | NF4 | | Authors: | Kobayashi, N, Nagashima, T, Minami, S, Koga, R, Chikenji, T, Koga, N. | | Deposit date: | 2020-03-24 | | Release date: | 2021-03-24 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Exploration of novel alpha-beta-protein folds through de novo design
Nat.Struct.Mol.Biol., 2023
|
|
7BQD
 
 | | Solution NMR structure of NF8 (knot fold); de novo designed protein with a novel fold | | Descriptor: | NF8 | | Authors: | Kobayashi, N, Sugiki, T, Fujiwara, T, Minami, S, Koga, R, Chikenji, G, Koga, N. | | Deposit date: | 2020-03-24 | | Release date: | 2021-03-24 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Exploration of novel alpha-beta-protein folds through de novo design
Nat.Struct.Mol.Biol., 2023
|
|
7BQE
 
 | | Solution NMR structure of NF3; de novo designed protein with a novel fold | | Descriptor: | NF3 | | Authors: | Kobayashi, N, Sugiki, T, Fujiwara, T, Minami, S, Koga, R, Chikenji, G, Koga, N. | | Deposit date: | 2020-03-24 | | Release date: | 2021-03-24 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Exploration of novel alpha-beta-protein folds through de novo design
Nat.Struct.Mol.Biol., 2023
|
|
7BQM
 
 | | Solution NMR structure of fold-0 Chantal; de novo designed protein with an asymmetric all-alpha topology | | Descriptor: | Chantal | | Authors: | Kobayashi, N, Sugiki, T, Fujiwara, T, Sakuma, K, Kosugi, T, Koga, R, Koga, N. | | Deposit date: | 2020-03-25 | | Release date: | 2021-04-07 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Design of complicated all-alpha protein structures
Nat.Struct.Mol.Biol., 2024
|
|
7BQN
 
 | | Solution NMR structure of fold-C Rei; de novo designed protein with an asymmetric all-alpha topology | | Descriptor: | Rei | | Authors: | Kobayashi, N, Sugiki, T, Fujiwara, T, Sakuma, K, Kosugi, T, Koga, R, Koga, N. | | Deposit date: | 2020-03-25 | | Release date: | 2021-04-07 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Design of complicated all-alpha protein structures
Nat.Struct.Mol.Biol., 2024
|
|
7BQQ
 
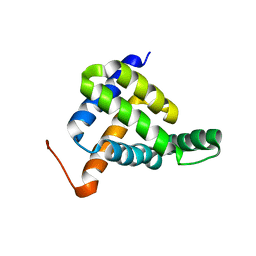 | | Solution NMR structure of fold-Z Gogy; de novo designed protein with an asymmetric all-alpha topology | | Descriptor: | Gogy | | Authors: | Kobayashi, N, Sugiki, T, Fujiwara, T, Sakuma, K, Kosugi, T, Koga, R, Koga, N. | | Deposit date: | 2020-03-25 | | Release date: | 2021-04-07 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Design of complicated all-alpha protein structures
Nat.Struct.Mol.Biol., 2024
|
|
7BQR
 
 | | Solution NMR structure of fold-K Mussoc; de novo designed protein with an asymmetric all-alpha topology | | Descriptor: | Mussoc | | Authors: | Kobayashi, N, Nagashima, T, Sakuma, K, Kosugi, T, Koga, R, Koga, N. | | Deposit date: | 2020-03-25 | | Release date: | 2021-04-07 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Design of complicated all-alpha protein structures
Nat.Struct.Mol.Biol., 2024
|
|
7BQS
 
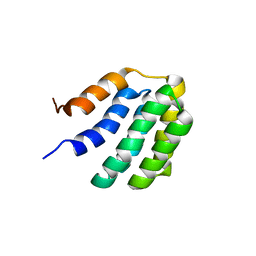 | | Solution NMR structure of fold-U Nomur; de novo designed protein with an asymmetric all-alpha topology | | Descriptor: | Nomur | | Authors: | Kobayashi, N, Nagashima, T, Sakuma, K, Kosugi, T, Koga, R, Koga, N. | | Deposit date: | 2020-03-25 | | Release date: | 2021-04-07 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Design of complicated all-alpha protein structures
Nat.Struct.Mol.Biol., 2024
|
|
7BUL
 
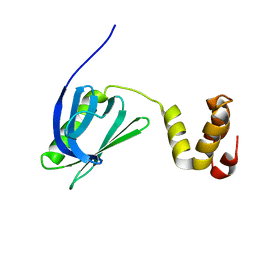 | |
7BUT
 
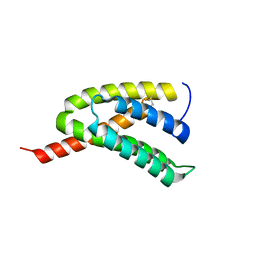 | |
7BV9
 
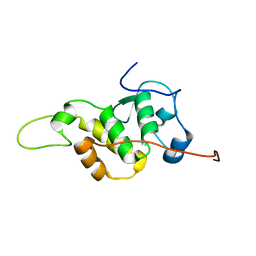 | | The NMR structure of the BEN domain from human NAC1 | | Descriptor: | Nucleus accumbens-associated protein 1 | | Authors: | Nagata, T, Kobayashi, N, Nakayama, N, Obayashi, E, Urano, T. | | Deposit date: | 2020-04-09 | | Release date: | 2021-02-17 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Nucleus Accumbens-Associated Protein 1 Binds DNA Directly through the BEN Domain in a Sequence-Specific Manner.
Biomedicines, 8, 2020
|
|
7BW5
 
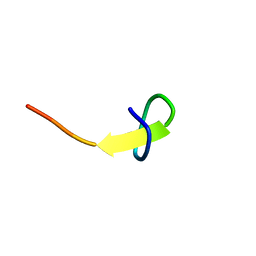 | | a new lasso peptide koreensin | | Descriptor: | lasso peptide koreensin | | Authors: | Hemmi, H, Kodani, S. | | Deposit date: | 2020-04-13 | | Release date: | 2020-09-09 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Heterologous production of new lasso peptide koreensin based on genome mining.
J Antibiot (Tokyo), 74, 2021
|
|
7BWE
 
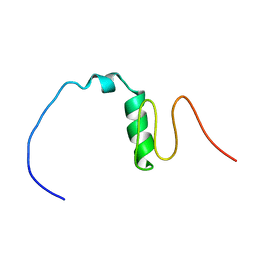 | | Consensus Chitin binding domain | | Descriptor: | Chitin binding beak protein 3 | | Authors: | Mohanram, H, Miserez, A. | | Deposit date: | 2020-04-14 | | Release date: | 2021-04-14 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Structure of a consensus chitin-binding domain revealed by solution NMR.
J.Struct.Biol., 213, 2021
|
|
7BWI
 
 | | Solution structure of recombinant APETx1 | | Descriptor: | Kappa-actitoxin-Ael2a | | Authors: | Matsumura, K, Kobayashi, N, Kurita, J, Nishimura, Y, Yokogawa, M, Imai, S, Shimada, I, Osawa, M. | | Deposit date: | 2020-04-14 | | Release date: | 2020-12-23 | | Last modified: | 2024-10-16 | | Method: | SOLUTION NMR | | Cite: | Mechanism of hERG inhibition by gating-modifier toxin, APETx1, deduced by functional characterization.
Bmc Mol Cell Biol, 22, 2021
|
|
7BWO
 
 | | Consensus chitin binding protein | | Descriptor: | Chitin binding beak protein 3 | | Authors: | Mohanram, H, Miserez, A. | | Deposit date: | 2020-04-15 | | Release date: | 2021-04-14 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Structure of a consensus chitin-binding domain revealed by solution NMR.
J.Struct.Biol., 213, 2021
|
|
7BX2
 
 | |
7BY7
 
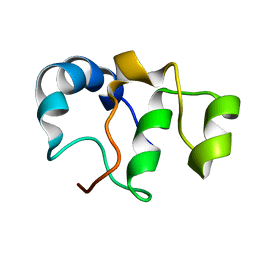 | | Bacteriophage SPO1 protein Gp46 | | Descriptor: | Putative gene 46 protein | | Authors: | Liu, B, Zhang, P. | | Deposit date: | 2020-04-22 | | Release date: | 2021-04-28 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Bacteriophage protein Gp46 is a cross-species inhibitor of nucleoid-associated HU proteins
Proc.Natl.Acad.Sci.USA, 119, 2022
|
|
7BZ7
 
 | | Template lasso peptide C24 mutant F15Y | | Descriptor: | lasso peptide | | Authors: | Liu, X.H, Liu, T, Ma, X.J, Yu, J.H, Yang, D.H, Ma, M. | | Deposit date: | 2020-04-27 | | Release date: | 2021-04-28 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Rational generation of lasso peptides based on biosynthetic gene mutations and site-selective chemical modifications.
Chem Sci, 12, 2021
|
|
7BZ8
 
 | | Template lasso peptide C24 mutant V3A | | Descriptor: | lasso peptide | | Authors: | Liu, X.H, Liu, T, Ma, X.J, Yu, J.H, Yang, D.H, Ma, M. | | Deposit date: | 2020-04-27 | | Release date: | 2021-04-28 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Rational generation of lasso peptides based on biosynthetic gene mutations and site-selective chemical modifications.
Chem Sci, 12, 2021
|
|
7BZ9
 
 | | Template lasso peptide C24 mutant I4A | | Descriptor: | lasso peptide | | Authors: | Liu, X.H, Liu, T, Ma, X.J, Yu, J.H, Yang, D.H, Ma, M. | | Deposit date: | 2020-04-27 | | Release date: | 2021-04-28 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Rational generation of lasso peptides based on biosynthetic gene mutations and site-selective chemical modifications.
Chem Sci, 12, 2021
|
|
7BZA
 
 | | Template lasso peptide C24 | | Descriptor: | lasso peptide | | Authors: | Liu, X.H, Liu, T, Ma, X.J, Yu, J.H, Yang, D.H, Ma, M. | | Deposit date: | 2020-04-27 | | Release date: | 2021-04-28 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Rational generation of lasso peptides based on biosynthetic gene mutations and site-selective chemical modifications.
Chem Sci, 12, 2021
|
|
7BZH
 
 | |
7C1M
 
 | | Complex structure of tyrosinated alpha-tubulin carboxy-terminal peptide and A1aY1 binder | | Descriptor: | Carboxy-terminal peptide from tyrosinated alpha-tubulin, Nanobody binder from SSO7d library | | Authors: | Kesarwani, S, Reddy, P.P, Sirajuddin, M, Das, R. | | Deposit date: | 2020-05-05 | | Release date: | 2020-09-02 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Genetically encoded live-cell sensor for tyrosinated microtubules.
J.Cell Biol., 219, 2020
|
|
7C36
 
 | | c-Myc DNA binding protein structure | | Descriptor: | RNA-binding motif, single-stranded-interacting protein 1 | | Authors: | Aggarwal, P, Bhavesh, N.S. | | Deposit date: | 2020-05-11 | | Release date: | 2021-05-12 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Hinge like domain motion facilitates human RBMS1 protein binding to proto-oncogene c-myc promoter.
Nucleic Acids Res., 49, 2021
|
|
