5WPX
 
 | |
5WPY
 
 | |
5WQ1
 
 | |
5WQZ
 
 | |
5WRX
 
 | | VG13P structure in LPS | | Descriptor: | analogue peptide VG13P | | Authors: | Datta, A, Bhunia, A. | | Deposit date: | 2016-12-04 | | Release date: | 2017-05-10 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Structural and Dynamic Insights into a Glycine-Mediated Short Analogue of a Designed Peptide in Lipopolysaccharide Micelles: Correlation Between Compact Structure and Anti-Endotoxin Activity.
Biochemistry, 56, 2017
|
|
5WT7
 
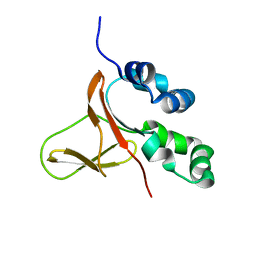 | | FAS1-IV domain of Human Periostin | | Descriptor: | Periostin | | Authors: | Yun, H, Lee, C.W. | | Deposit date: | 2016-12-09 | | Release date: | 2017-12-13 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | 1H,13C, and 15N resonance assignments of FAS1-IV domain of human periostin, a component of extracellular matrix proteins.
Biomol NMR Assign, 12, 2018
|
|
5WUZ
 
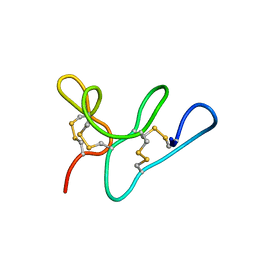 | | Morintides mO1 | | Descriptor: | Morintide mO1 | | Authors: | Xiao, T, Tam, J.P. | | Deposit date: | 2016-12-21 | | Release date: | 2017-01-25 | | Last modified: | 2024-11-13 | | Method: | SOLUTION NMR | | Cite: | Morintides: 8C-Hevein-like peptides without a protein cargo from the drumstick tree Moringa oleifera
To Be Published
|
|
5WXE
 
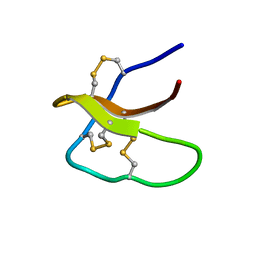 | | Highly disulfide-constrained antifeedant jasmintides from Jasminum sambac flowers | | Descriptor: | jasmintide js3 | | Authors: | Kumari, G, Wong, K.H, Serra, A, Shin, J, Yoon, H.S, Sze, S.K, James, P.T. | | Deposit date: | 2017-01-07 | | Release date: | 2018-01-17 | | Last modified: | 2024-11-20 | | Method: | SOLUTION NMR | | Cite: | Highly disulfide-constrained antifeedant jasmintides from Jasminum sambac flowers
To Be Published
|
|
5WYE
 
 | |
5WYO
 
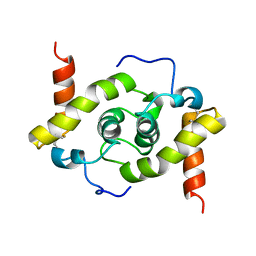 | | Solution structure of E.coli HdeA | | Descriptor: | Acid stress chaperone HdeA | | Authors: | Yang, C, Hu, Y, Jin, C. | | Deposit date: | 2017-01-14 | | Release date: | 2017-11-22 | | Last modified: | 2024-10-09 | | Method: | SOLUTION NMR | | Cite: | Characterizations of the Interactions between Escherichia coli Periplasmic Chaperone HdeA and Its Native Substrates during Acid Stress
Biochemistry, 56, 2017
|
|
5X0S
 
 | | Solution NMR structure of peptide toxin SsTx from Scolopendra subspinipes mutilans | | Descriptor: | SsTx | | Authors: | Wu, F, Luo, L, Qu, D, Zhang, L, Tian, C, Lai, R. | | Deposit date: | 2017-01-23 | | Release date: | 2018-01-24 | | Last modified: | 2024-10-23 | | Method: | SOLUTION NMR | | Cite: | Centipedes subdue giant prey by blocking KCNQ channels
Proc. Natl. Acad. Sci. U.S.A., 115, 2018
|
|
5X1X
 
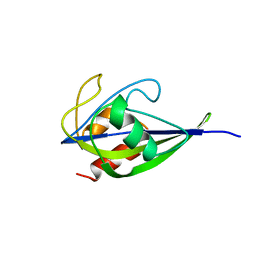 | |
5X29
 
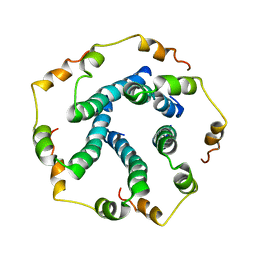 | |
5X34
 
 | |
5X35
 
 | |
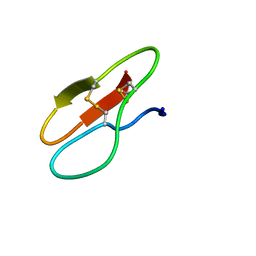
5X36
 
 | |
5X37
 
 | |
5X38
 
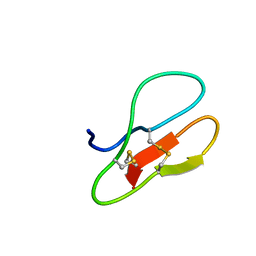 | |
5X39
 
 | |
5X3C
 
 | |
5X3L
 
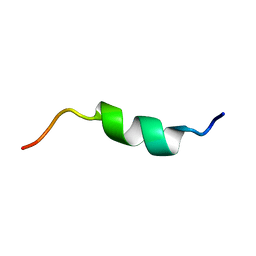 | |
5X3Y
 
 | | Refined solution structure of musashi1 RBD2 | | Descriptor: | RNA-binding protein Musashi homolog 1 | | Authors: | Iwaoka, R, Nagata, T, Tsuda, K, Imai, T, Okano, H, Kobayashi, N, Katahira, M. | | Deposit date: | 2017-02-09 | | Release date: | 2017-12-13 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Structural Insight into the Recognition of r(UAG) by Musashi-1 RBD2, and Construction of a Model of Musashi-1 RBD1-2 Bound to the Minimum Target RNA
Molecules, 22, 2017
|
|
5X3Z
 
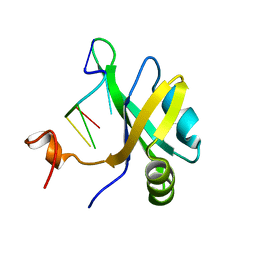 | | Solution structure of musashi1 RBD2 in complex with RNA | | Descriptor: | RNA (5'-R(*GP*UP*AP*GP*U)-3'), RNA-binding protein Musashi homolog 1 | | Authors: | Iwaoka, R, Nagata, T, Tsuda, K, Imai, T, Okano, H, Kobayashi, N, Katahira, M. | | Deposit date: | 2017-02-09 | | Release date: | 2017-12-13 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Structural Insight into the Recognition of r(UAG) by Musashi-1 RBD2, and Construction of a Model of Musashi-1 RBD1-2 Bound to the Minimum Target RNA
Molecules, 22, 2017
|
|
5X4F
 
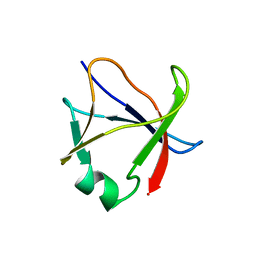 | |
5X5S
 
 | |
